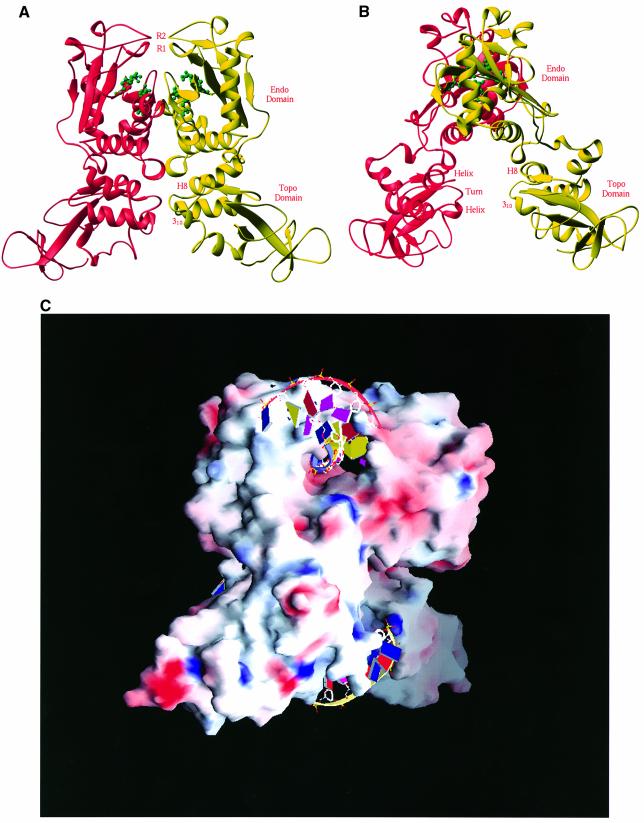
Fig. 2. Structure of the NaeI dimer. (A) Two monomers in red and yellow ribbons are related by a molecular 2-fold axis located vertically. The tentative active site residues of Glu70, Asp86, Asp95 and Lys97 for the endonuclease activity are shown in green ball-and-stick representation. Two putative recognition loops are located at the very top of the picture, and are marked R1 (residues 102–109, β-strand B4) and R2 (residues 141–149, a β-like turn). (B) A view of the NaeI dimer with a 90° rotation about the vertical axis from (A). The CAP DNA-binding motif in the Topo domain of NaeI, labeled helix–turn–helix and H8 and 310 (before H9), is common for many of the DNA-binding proteins such as topoisomerases and transcription factors. This site is presumably a second site of DNA binding and responsible for the topoisomerase activity of the NaeI L43K mutant. (C) Surface presentation of dimeric NaeI: red for negatively charged residues and blue for positively charged residues. Two DNA fragments are modeled into the endonuclease (top) and topoisomerase (bottom) sites on the basis of the superposition of NaeI over the PvuII–DNA and CAP–DNA structures, respectively.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.