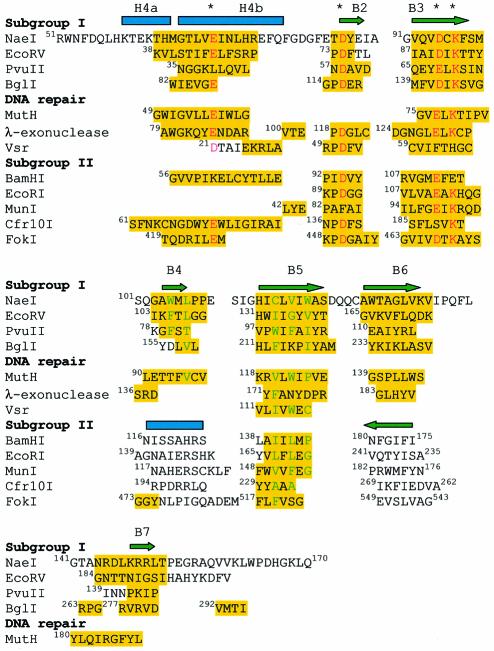
Fig. 4. Structure-based sequence alignment of the NaeI Endo domain with partial sequences of eight restriction endonucleases, two DNA repair endonucleases and λ-exonuclease. Amino acids highlighted in yellow are used in the calculation of the r.m.s. deviation of the superposition. Three negatively charged residues for divalent metal binding and the catalytic lysine are marked with asterisks and colored red. Hydrophobic residues of the molecule core are colored green. The alignment suggests that the restriction endonucleases are divided into subgroup I (NaeI, EcoRV, PvuII and BglI) and subgroup II (BamHI, EcoRI, MunI, Cfr10I and FokI). Subgroup II shows a β-strand running in an opposite direction to B6 in NaeI, and also a helix in place of β-strand B4 in NaeI. B4 and B7 in NaeI are, respectively, contained in recognition loops R1 and R2 (Figure 2A).

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.