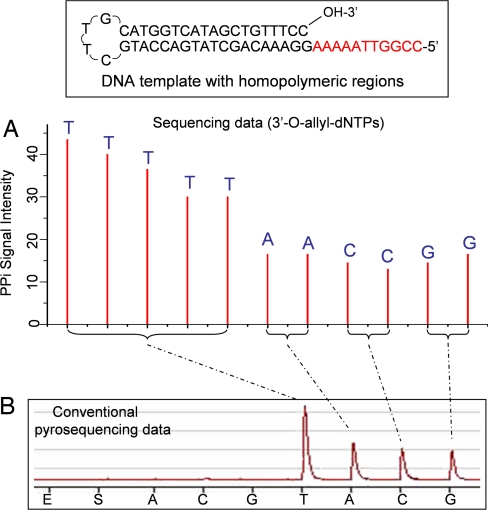
Fig. 6.
Comparison of reversible terminator-pyrosequencing using 3′-O-allyl-dNTPs with conventional pyrosequencing using natural nucleotides. (A) The self-priming DNA template with stretches of homopolymeric regions (five A, two T, two G, and two C bases) was sequenced by using 3′-O-allyl-dNTPs. The homopolymeric regions are clearly identified, with each peak corresponding to the identity of each base in the DNA template. (B) Pyrosequencing data using natural nucleotides. The homopolymeric regions produced one large peak corresponding to the stretch of T bases and three smaller peaks for stretches of A, C, and G bases. However, it is very difficult to decipher the exact sequence from the data.