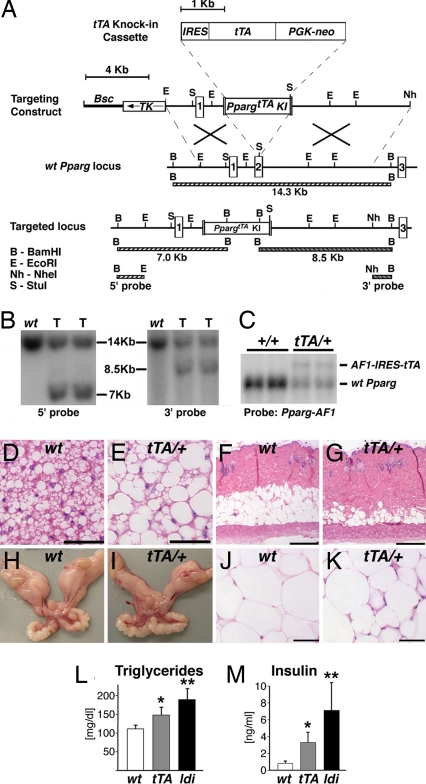
Fig. 4.
tTA causes mild lipodystrophy with modest metabolic anomalies. (A) The PpargtTA allele contains an exact replica of the IRES, tTA, Pgk-neo, and synthetic polyA modules of the Ppargldi allele, targeted to the same location within the second coding exon of the Pparg gene. (B) Southern blots of BamHI-digested genomic DNA hybridized to 5′ and 3′ external probes. Correctly targeted PpargtTA/+ ES cell clones (T) exhibit the digestion patterns predicted in A. (C) A Northern blot of two WT (+/+) and two PpargtTA/+ EFPs was hybridized the 5′ Pparg cDNA probe, as described in Fig. 1C. PpargtTA/+ WAT expresses the same AF1-IRES-tTA fusion transcript seen in Ppargldi/+ fat. (D–K) BAT (D and E), skin (F and G), EFPs (H and I), and WAT (J and K) from 10-wk-old WT (D, F, H, and J) and PpargtTA/+ (E, G, I, and K) males. BAT exhibits approximately the same phenotype of Ppargldi/+ mice (see Fig. 2C), whereas subdermal fat and WAT exhibit a markedly attenuated phenotype (compare with Fig. 2 D, I, and J. (Scale bars: D, E, J, and K, 50 μm; F and G, 250 μm.) (L and M) Fasting triglyceride (L) and insulin (M) levels in 5.5-wk-old WT, PpargtTA/+, and Ppargldi/+ males (n = 5–6 mice). All cohorts were born within the same week and assayed simultaneously. Notice the intermediate hypertriglyceridemic and hyperinsulinemic phenotypes of PpargtTA/+ compared with Ppargldi/+ mice. * and **, levels of both analytes are significantly different in each strain compared with the other two (P < 0.05).