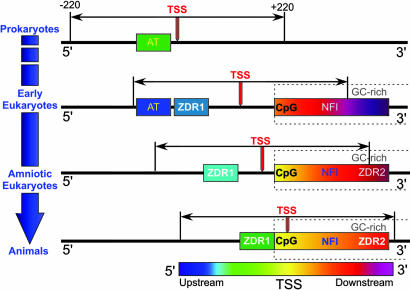
Fig. 4.
Model for the emergence of GC-rich transcriptional elements and migration of the transcription start site (TSS, red arrow) of genes from prokaryotes to early eukaryotes to amniotic eukaryotes, and, finally, to higher eukaryotes. In this model, the prokaryotic AT-rich promoters and the GC-rich eukaryotic elements are seen to be fixed, whereas the TSS and the analysis window (±220 bp relative to the TSS) migrate in the 3′ direction as the size and complexity of the transcriptosome increases. In prokaryotes, the primary transcriptional control elements are AT-rich upstream promoters, which account for the strong suppression of GC-rich elements that are characteristic of eukaryotes. Early eukaryotes show both the persistence of localized upstream AT-rich promoters that are characteristic of prokaryotes, as well as the accumulation of the eukaryotic GC-rich elements (CpG and NFI elements) as the GC content (dashed boxes) downstream of the TSS increases. The first Z-DNA regions (ZDR1) also emerge at this point, independently of the other GC-rich elements. In progressing toward amniotic eukaryotes, the TSS migrates further in the 3′ direction, followed by the ZDR1 elements. This is in concert with AT-rich promoters becoming less distinct (and, therefore, their locations are not specified in this figure), the emergence of a second class of CG-rich Z-DNAs (ZDR2) accumulated downstream of the TSS, and convergence of the upstream ZDR1 sites with the GC-rich elements.