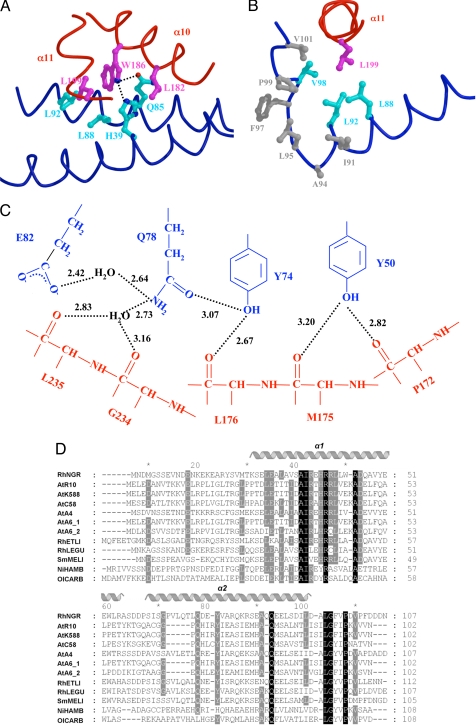
Fig. 3.
Structural analysis of the TraRNGR–TraMNGR complex. (A) The TraRNGR–TraMNGR interactions at TraRNGR W186. Side chains from TraRNGR are colored in magenta, and those from TraMNGR are in cyan. The oxygen atom is in red, and nitrogen atoms are in blue. (B) Interactions of TraRNGR L199 (in magenta) with the hydrophobic cluster at the C termini of TraMNGR. Hydrophobic side chains of the cluster are colored in gray, whereas those that interact directly with L199 are colored in cyan. (C) The extensive intermolecular hydrogen-bonding network. Residues from TraMNGR and TraRNGR are colored in blue and red, respectively. The values (in angstroms) denote the distance between hydrogen donors and acceptors. Only interacting moieties, i.e., side chains of TraMNGR and backbones of TraRNGR, are shown. (D) Multiple sequence alignment of the antiactivator TraM proteins by using CLUSTALW (30). Amino acid sequences are from the following bacteria: RhNGR (or TraMNGR in the text), Rhizobium sp. NGR234; AtR10 (TraMAt), A. tumefaciens R10; AtK588, A. tumefaciens K588; ATC58, A. tumefaciens C58; AtA4, A. tumefaciens A4; AtA_1, A. tumefaciens A6; AtA_2, A. tumefaciens A6 traM2; RhETLI, Rhizobium etli CFN42; RhLEGU, R. leguminosarum; SmMELI, Sinorhizobium meliloti AK631; and NiHAMB, Nitrobacter hamburgensis X14; OlCARB, Oligotropha carboxidovorans. Invariant residues are highlighted in black, and highly conserved resides are shaded in grey. Secondary structure elements of TraMNGR are indicated above. A and B were generated by using MOLSCRIPT and RASTER 3D (28, 29).