Figure 1.
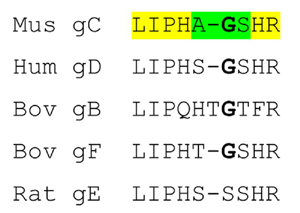
(a) Motif alignment of selected γ-crystallins.
The sequence of human γC-crystallin [GI:10518338] is aligned with other known mammalian γC-crystallins (bovine [GI:600873], rhesus monkey [EF426305], guinea pig [EF426306], dog [EF426309], C57Bl/6 mouse [EF426301] and C57Bl/6 mouse γCins [EF426302], and with some other members of the γ-crystallin family (human γA [EF426311], human γB [GI:117461], bovine γB [GI:162918], C57Bl/6 mouse γB [EF426303], rhesus monkey γB [EF426304], guinea pig γB [EF426307] and dog γB [EF426308]. The two motifs from the N-terminal domain are aligned above the two from the C-terminal domain. Residues referred to in the text are highlighted. The residues highlighted in red and blue in each motif form an inter-motif ion pair, with the acidic residue from one motif closely associated with the basic residue in the other. Both the basic residues on the N-terminal domain are altered in proteins that have solubility problems: in wild-type human γC-crystallin the motif 2 Arg is replaced by Cys and in a human γD-crystallin cataract mutant, the motif 1Arg is Ser. Both these changes disrupt the tight ion pair and leave an unpartnered acidic side chain. The occurrence of an amide containing side chain in motif 3 does not seem to affect the interaction, with the amide group hydrogen bonding to the acidic side chain in motif 4.
Abbreviations: Hum - Human, Mus - Mouse, Rh - Rhesus Monkey, GP - Guinea Pig, Bov - Bovine.

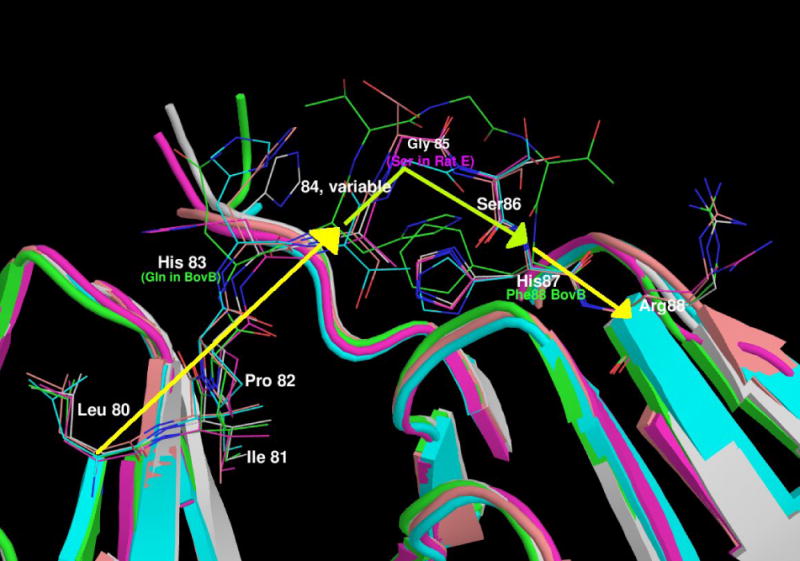
(b) A view of the linkers of all the γ-crystallins with solved structures.
The domains are shown in cartoon format, N-terminal domain to the left and the C-terminal domain on the right with the intervening residues shown in stick format. The proteins are coloured as follows: Mouse γC in pink, human γD in grey, bovine γB in green, bovine γF in turquoise and rat γE in orange. It can be seen that the relatively conserved LIPHX section of the intervening sequence continues from β-strand D2 of the N-terminal domain, whilst the SHR section continues into β-strand A3 in the C-terminal domain. In all the proteins, the consensus G (S in the case of Rat γE) separates the two ‘straight’ parts of the linker with a sharp turn. The presence of the extra T in bovine γB pushes the C-terminal part of the linker away, but in all the proteins it is the consensus glycine where the linker takes a sharp turn. Therefore, structurally the gap in the shorter sequences lies in the N-terminal end of the linker before the location of the turn.