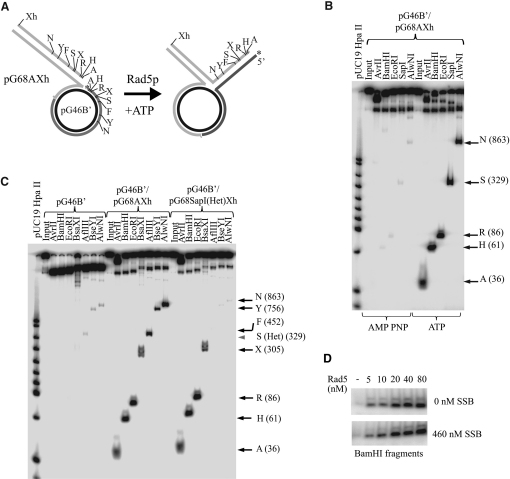
Figure 3.
Rad5 Can Regress Plasmid-Sized Model Forks
(A) Schematic representation of the joint DNA substrate (pG46B′/pG68AXh) and the outcome of its Rad5-mediated regression. Letters A, H, R, X, S, F, Y, N, and Xh refer to restriction endonuclease sites AvrII, BamHI, EcoRI, BsaXI, SapI, AflIII, BseYI, AlwNI, and XhoI, respectively. The positions of 5′ 32P labels on the “lagging strand” are marked with asterisk.
(B) The extent of Rad5-dependent fork regression. The restriction enzyme site transfer to the regressed arm by Rad5 was followed in the presence of 5 mM ATP/Mg. The positions of the various restriction products generated by digestion of the regressed fork are indicated. Reaction with 5 mM AMP-PNP/Mg shows the background level of spontaneous regression.
(C) Fork regression by Rad5 is progressive. Fork regression by Rad5 was compared on two joint DNA substrates containing either no heterology, or in which a 30 base pair sequence heterology was introduced at the SapI site, shown by arrowhead, of pG68A (named pG68 SapI[Het]). We note that the regression beyond the heterology was blocked, as revealed by the absence of F-, Y-, and N-specific bands. Reaction with gapped DNA (pG46B′) shows background due to nicking at the gap.
(D) Fork reversal is not affected by E. coli ssDNA-binding protein. Regression through the BamHI site was monitored at various Rad5 concentrations (0–80 nM) in the presence or absence of E. coli SSB protein (460 nM).