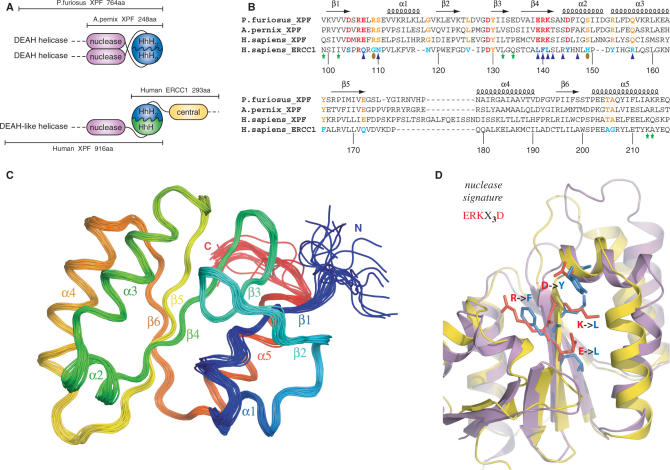
Figure 1.
(A) Domain organization of the archaeal XPF homodimeric members and the human ERCC1/XPF heterodimer. (B) Structure-based sequence alignment of the nuclease XPF domains from archaea to human and the corresponding central domain of human ERCC1. Secondary structure elements of the prototype XPF nuclease fold are indicated at the top of the sequences. Catalytic residues in the XPF nucleases are colored red and their corresponding substitutions in ERCC1 blue. Other invariant residues in XPF domains are depicted in orange, while the ERCC1 equivalents are depicted in cyan. Residues of cERCC1 perturbed largely upon XPA titration are indicated by blue triangles and those appear only in the final complex by brown ellipses. Green asterisks indicate cERCC1 residues perturbed by DNA titration. cERCC1 sequence is numbered at the bottom. (C) Ensemble of the final 20 structural conformers of cERCC1 as determined by solution NMR. Secondary structure elements and N- and C-termini are labeled. (D) Superposition of the crystal XPF nuclease structure (2bgw) from A. pernix (purple) and the solution NMR structure (2jpd) of human cERCC1 (yellow). Emphasis is given to the nuclease signature and the corresponding substitutions in cERCC1.