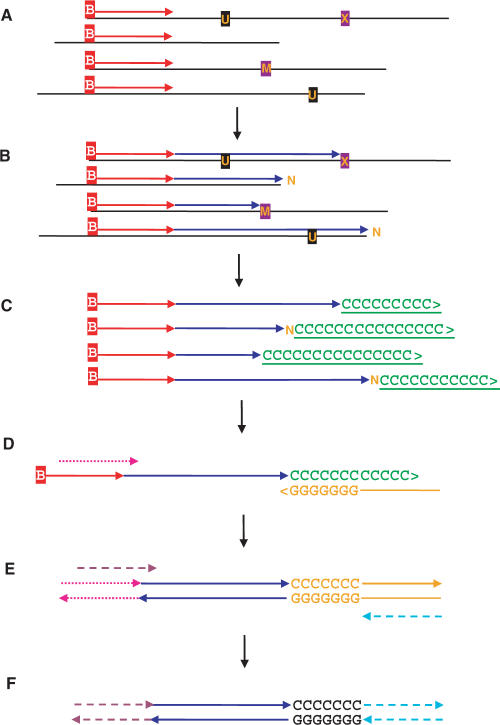
Figure 1.
Single primer extension (SPEX) amplification. (A) Denaturation and hybridization of a single biotinylated primer to one target strand at the locus-of-interest. (B) Primer extension by a thermostable DNA polymerase until halted at the physical end of an aDNA template molecule; at a polymerase-blocking modified base [M]; or at an abasic site, some other non-coding lesion, or some kind of physical block [X]. Miscoding lesions [U] do not block primer extension, but result in altered sequences. Polymerases can catalyze the non-directed addition of a single 3′-terminal nucleotide [N] following primer extension to either the physical end of a fragmented aDNA template, or to an abasic site or other non-coding lesion. Single or multiple cycles of SPEX primer extension can be used. Biotinylated molecules were then bound to Streptavidin-coated beads and stringent washes removed everything else (e.g. aDNA template molecules, enzymes, buffer, etc). (C) Biotinylated primers and extended primers (with single-stranded, direct first-generation copies of individual aDNA template molecules) were then polyC-tailed using terminal transferase (TdT), followed again by bead-wash removal of TdT and buffer. (D) Locus-specific, primer-extended, polyC-tailed ssDNA molecules were then selectively amplified by PCR; using a partially-nested, locus-specific, SPEX-2 forward primer (Tables S1 and S2) and a polyG-based, 5′ adapter-tagged, reverse primer (Tables S1 and S2). (E) These products were amplified a final time by PCR using a further partially nested, locus-specific, SPEX-3 forward primer (Tables S1 and S2) and a 5′-adapter reverse primer (Tables S1 and S2). (F) The final products of SPEX amplification underwent restriction digestion, directional cloning and sequencing.