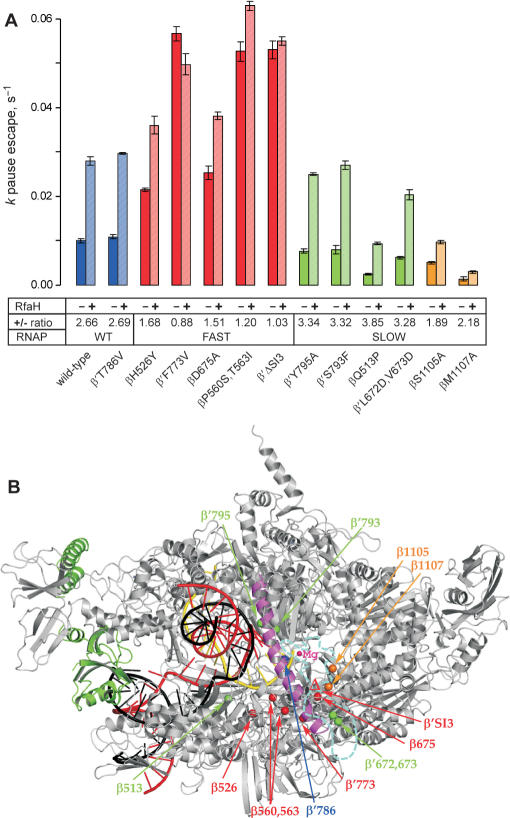
Figure 3.
The summary of the RfaH effects on pausing at the hisP site by the RNAP variants. (A). The data from experiments similar to those shown in Figures 2B were analyzed as described in (20); for each enzyme, the assay was repeated at least twice. The kp for escape from the pause was measured in the presence and in the absence of RfaH; the ratio is presented below each variant. The pause efficiency cannot be determined reliably, particularly for the slow enzymes which display significant asynchrony during elongation. The substitutions that are fast and resistant to RfaH are colored in red, those that displays the wild-type elongation rate and response to RfaH are colored in blue, the substitutions that are slow and hypersensitive to RfaH—in green. Finally, the β S1105 and β M1107 residues, whose substitutions to Ala both reduce the elongation rate and the response to RfaH, are shown in orange. (B). A model of RfaH bound to the TEC. The Thermus thermophilus RNAP (46) is shown in gray with the bridge helix highlighted in magenta, the template DNA—in red, the NT DNA—in black, the nascent RNA—in yellow. Position of the RNAP active site is marked by the high-affinity catalytic Mg2+ ion (small magenta sphere). RfaH (green) is bound to the β′ CH. The Cα atoms of the RNAP substitutions tested in this work are shown as spheres and colored according to their response to RfaH as in panel A. The disordered β′ trigger loop is depicted as a cyan dashed line, with the β′SI3 domain (missing in T. thermophilus) inserted in the middle; the βΔ′SI3 deletion variant used here lacks E. coli β′ residues 943–1130 (6). The distance between the RfaH contact site on the β′ CH and the nearest residue shown (β Q513) is 53 Å.