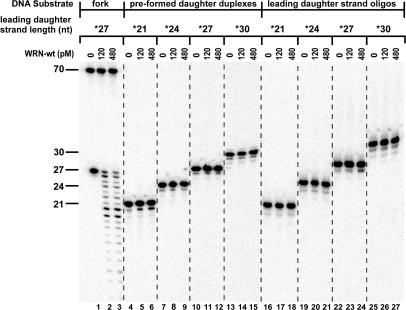
Figure 3.
Comparison of WRN exonuclease activity on substrates and products of fork regression reactions. WRN-wt (0, 120 or 480 pM) was incubated at 37°C for 5 min with 50 pM of fork substrate containing a leading arm gap of 5 nt [*27lead/70lead-*70lag/30lag (lanes 1–3)], pre-formed daughter duplex substrates [*21lead/30lag (lanes 4–6)], *24lead/30lag (lanes 7–9), *27lead/30lag (lanes 10–12) and *30lead/30lag (lanes 13–15)] or isolated leading daughter strand oligomers [*21lead (lanes 16–18), *24lead (lanes 19–21), *27lead (lanes 22–24) and *30lead (lanes 25–27)], with the asterisks above indicating the radiolabeled strands. The resulting DNA species were analyzed by denaturing PAGE and phosphorimaging, with the lengths (in nt) and positions of migration of the labeled, undigested oligomers within various substrates indicated at left. Importantly, WRN exonuclease activity is only detectable on the leading daughter strand in the context of the intact fork substrate.