Abstract
Background:
We hypothesized that acetylation of the Stat1 regulates interferon-γ (IFN-γ) mediated macrophage expression of inducible nitric oxide synthase (iNOS).
Methods:
RAW 264.7 iNOS expression was induced with IFN-γ. Deacetylase inhibitors trichostatin A (TSA) or valproic acid (VPA) were added. Stat1 and iNOS mRNA and protein were measured. Acetylated Stat1 was determined by immunoprecipitation. Chromatin immunoprecipitation assessed in vivo binding of Stat1 to the iNOS promoter.
Results:
IFN-γ significantly increased nitrite, iNOS protein and iNOS mRNA, and iNOS promoter activation. (p<0.01 vs Control for nitrite, protein and mRNA). TSA mediated acetylation decreased these to levels that were not different from Controls. IFN-γ increased acetylated Stat1 by five-fold (p<0.02 vs. Control); TSA+IFN-γ caused an additional 4-fold increase in acetylated Stat1 (p<0.05 vs IFN alone). Stat1 binding to the iNOS promoter increased 8-fold with IFN-γ (p<0.01 vs Control) In TSA+IFN-γ, Stat1 binding was not different from Controls. Though less potent than TSA, VPA also significantly decreased nitrite, iNOS protein, iNOS mRNA, Stat1 acetylation and Stat1 binding.
Conclusions:
Acetylation of Stat1 protein correlates with decreased Stat1 binding to the iNOS promoter with resultant inhibition of IFN-γ mediated iNOS expression. Acetylation of the Stat1 protein may down regulate iNOS expression in pro-inflammatory states.
Introduction
In sepsis, pro-inflammatory cytokines are elaborated, and iNOS is systemically expressed in multiple cell types, including macrophages.(1) The sustained production of NO in high concentration regulates multiple cellular and biochemical functions, including inotropic and chronotropic cardiac responses, systemic vasomotor tone, intestinal epithelial permeability, endothelial activation, and microvascular permeability. (2-4) An essential role for the Stat1 pathway in iNOS induction had been demonstrated for murine, rat and human cells. All mammalian iNOS promoters contain several homologies with the IFN-γ-regulated transcription factor Stat1α binding sites (GAS).(5) However, while the molecular pathways which upregulate iNOS expression have been extensively studied in multiple cell types, including the macrophage, little is known of the parallel counter-regulatory pathways which repress or inhibit macrophage iNOS expression in this context.
Post-translational modification of proteins with resultant alteration in function is a well established regulatory mechanism in cell biology. Recently, it has become evident that regulated acetylation of nonhistone proteins may significantly alter cellular activities. Nonhistone protein acetylation may be as pervasive and important as phosphorylation.(6) The equilibrium of nonhistone protein acetylation and deacetylation is maintained by histone acetyltransferases (HATs) and histone deacetylases (HDACs) and is a dynamic regulatory system modulating protein function.(6) In this regard, we investigated the role of acetylation of Stat1 in regulation of IFN-γ mediated iNOS expression in RAW264.7 murine macrophages. Our results indicate that Stat1 acetylation is associated with decreased Stat1 binding to the iNOS promoter and decreased iNOS expression. This has not been previously described and suggests that acetylation of Stat1 may serve to down regulate macrophage iNOS expression.
Methods
Cell culture. RAW 264.7 macrophages were maintained in DMEM with 10% heat-inactivated FCS, 100 U/ml penicillin, and 100 µg/ml streptomycin, and incubated in 5% CO2-95% air at 37°C. IFN-γ (500 u) was used to induce NO synthesis. In selected instances, the deacetylase inhibitors trichostatin A (TSA; 200 nM) and valproic acid (VPA; 1.5 mM) were used. After incubation for 3, 12, and/or 24 hr, the supernatants and cells were harvested. Assay of NO production. After stimulation, 50 µl of culture medium was mixed with 50 µl of 1% sulfanilamide dissolved in 0.5 mol/L HCl. After 5-min incubation at room temperature, 50 µl of N-(1-naphthyl)-ethylenediamine was added. Following incubation for 10 min at room temperature, the absorbance of samples was measured at 540 nm and compared with NaNO3 standards. RNA preparation and RT-PCR. Total RNA was isolated from RAW 264.7 macrophages using TRIzol reagent (Life Technologies, Rockville, MD). Equal amounts of mRNA (0.5 µg) were reverse-transcribed into cDNA using the iScript cDNA synthesis kit (Bio-Rad) according to manufacturer's instructions. The cDNA were used in subsequent PCR reactions; the primers for iNOS were 5′-CATCCATGCAAAGAACGTGT-3′ (forward) and 5′-GAAGGTGAGCTG AACGAGGA-3′ (reverse), and the primers for Stat1 were 5′-CTTATTCCATGGA CAAGGTTTTG-3′ (forward) and 5′-GGTGCTTCTTAATGAGCTCTAGG-3′ (reverse). Densitometric analysis was performed with AlphaImager™ 3400 to quantify the RT-PCR results. The intensities of the PCR products were normalized to that of housekeeping gene β-actin. Transient transfection analysis. 1×106 cells were plated on a 12-well plate and allowed to grow for 24 h before the transfection. 2 µg plasmid DNA and 2µg protamine sulfate diluted in OPTI-DMEM and 24 ug lipofectamine diluted in OPTI-DMEM were combined and incubated at room temperature for 20 min. Cells with transfection reagents were incubated for 4 h at 37 °C in a CO2 incubator. Transfection medium was then replaced with DMEM containing 10% FBS. At least 24 h later, the medium was changed, and cells were treated, as described. To control transfection efficiency between groups, 0.1 ug pRL-TK was added to each well. Cells were harvested in 0.4 ml of reporter lysis buffer (Promega, Madison, WI), and dual luciferase reporter assays were performed by following the protocol provided by the manufacturer. Western blotting. Cells were lysed in buffer (0.8% NaCl, 0.02 KCl, 1% SDS, 10% Triton X-100, 0.5% sodium deoxycholic acid, 0.144% Na2HPO4 and 0.024% KH2PO4, pH 7.4), put on ice for 30 min, homogenized, and and centrifuged at 12,000 x g for 15 min at 4 °C. The cell lysates were separated by 12% SDS-PAGE and transferred to polyvinylidene difluoride (PVDF) membrane and blocked by 5% dry milk PBST. Blocked membranes were then incubated with iNOS (1:1000; Transduction Laboratories), β-actin (1:1000; Cell Signaling, Danvers, MA), Stat1 p84/p91 (1:1000; Santa Cruz Biotechnology) or Tyr 701 Stat1 (1:1000; Santa Cruz Biotechnology) antibody for 1 hr at room temperature. After washing three times with PBST, the membranes were incubated with donkey anti-rabbit IgG-horse radish peroxidase (1:1000; Santa Cruz Biotechnology) for 1 hr at room temperature. After an additional three washes, protein-antibody complexes were visualized using Western Blotting Luminol Reagent (Santa Cruz Biotechnology). Co-immunoprecipitation (Co-IP). 750 µg of extracted protein were mixed with 500 µl of PBS, 1 µl of normal mouse IgG and 20 µl of G-plus agarose. Samples were rotated for 30min at 4°C. After centrifugation for 1 min at 2500 rpm at 4°C, anti-acetylated-lysine (Cell Signaling, Danvers, MA) antibody was added. Samples were rotated for 30 min again at 4°C. G-plus agarose was added to the samples, and they were rotated at 4°C overnight. After the beads were washed three times with PBS, the pellets were dissolved into NuPAGE® loading buffer (Invitrogen) after centrifugation. Protein was analyzed by western blotting with Stat1 p84/p91 (1:1000; Santa Cruz Biotechnology) antibody. Chromatin immunoprecipitation (ChIP)-Real Time PCR for NF-κB and Stat1 binding to iNOS promoter. Chromatin from macrophages was fixed and immunoprecipitated using the ChIP assay kit (Upstate Biotechnology, Lake Placid, NY). Sequences were identified for the mouse iNOS promoter (Genbank no. L09126). Purified chromatin was immunoprecipitated using 10 µg of anti-NF-κB p65 (Santa Cruz Biotechnology, Santa Cruz, CA), anti-Stat1 (Santa Cruz Biotechnology, Santa Cruz, CA) or 5 µl of rabbit nonimmune serum; eluted DNA fragments were purified to serve as templates. The input fraction corresponded to 0.1 and 0.05% of the chromatin solution before immunoprecipitation. The average size of the sonicated DNA fragments subjected to immunoprecipitation was 500 bp as determined by ethidium bromide gel electrophoresis. After DNA purification, the presence of the selected DNA sequence was quantified by RT-PCR. ChIP assays addressing NF-kB-1 utilized the PCR primers TGTACCTTAGACAAGGCAAAACA and TGAGTTCTAGGACAAACTAGGGCT, resulting in a 260 bp fragment, NF-kB-2 utilized AAACTGCAAATGAGAGAACAGACAG and TGTTTTGCATAAAGTCACTGTTCC, resulting in a 312 bp fragment and GAS utilized primers ACACGAGGCTGAGCTGACTT and CACACATGGCATGGAATTTT resulting in a 186 bp fragment. Statistical analysis. All data are presented as mean ± SEM of three or four experiments. Analysis was performed using a Students t test. Values of p < 0.05 were considered significant.
Results
RAW264.7 murine macrophages were exposed to IFN-γ (500 U/ml) for 0, 3, 12 and 24 hours. Unstimulated cells served as Controls. TSA and VPA were added in the presence and absence of IFN-γ to inhibit de-acetylase activity. NO was measured as nitrite, the NO metabolite. (Figure 1a) In the presence of IFN-γ, NO production was readily detected after 12 and 24 hours with concentrations of 62 ± 2.3 µM and 88 ± 4.8 µM, respectively. (p<0.01 vs Control at 12 hrs and 24 hrs) In IFN + TSA cells, NO production was completely ablated and was not different from that of unstimulated Controls at all time points. In IFN + VPA cells, NO production was present at 12 and 24 hours, albeit significantly less than that found in the presence of IFN alone. (p<0.03 vs IFN and Controls at 12 hrs and 24 hrs) Western blot analysis was performed for iNOS and Stat1 protein expression after 6 hours of IFN stimulation. (Figure 1b) Expression of iNOS was detected in IFN treated cells; in the presence of IFN + VPA, iNOS expression was 1/10 that found in the presence of IFN alone.(p<0.02) No iNOS protein was found in the presence of IFN + TSA. Total cellular Stat1 protein was unchanged under all stimulation conditions. Levels of iNOS and Stat1 mRNA at 6 hours were measured using reverse transcriptase PCR. (Figure 1c) The findings parallel those found for protein expression. iNOS mRNA was readily detected in IFN treated cells; in the presence of IFN + VPA, iNOS expression was significantly less than that found in the presence of IFN alone.(p<0.02) iNOS mRNA was not found in the presence of IFN + TSA. Stat1 mRNA was unchanged under all stimulation conditions. Finally, transient transfection assays with the murine iNOS promoter were carried out to determine effect of de-acetylase inhibition on iNOS promoter activation. (Figure 1d) The murine iNOS promoter contains 2 NF-κB (NF-κB-1 at position −114 to −104 and NF-κB-2 at position −1044 to −1034) and 1 GAS (position −934 to −942 bp) binding sites thought to be essential for iNOS gene transcription depending upon the stimulatory agent. IFN stimulation was associated with a 30-fold increase in luciferase activity in comparison to unstimulated Control cells. (p<0.001) In contrast, minimal iNOS promoter activity was found in IFN + TSA cells. Again, IFN + VPA was associated with increased iNOS promoter activation, but to an extent both significantly less than IFN alone and more than Control. (p<0.01 vs Control and IFN) These results indicate that global inhibition of de-acetylase activity significantly decreases IFN-γ mediated iNOS gene expression.
Figure 1. Acetylation and NO synthesis in IFN-γ stimulated RAW264.7 murine macrophages.
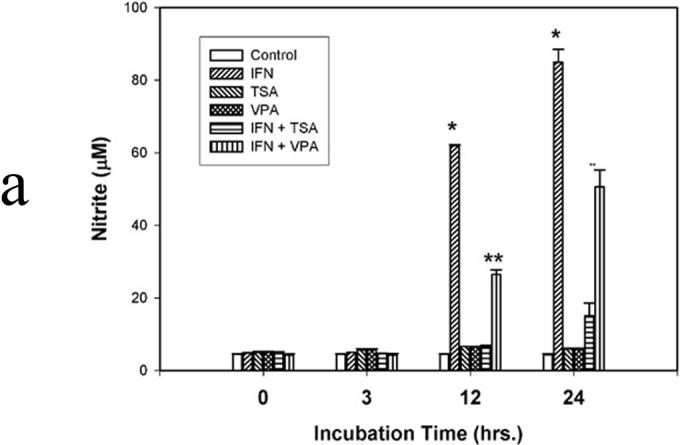
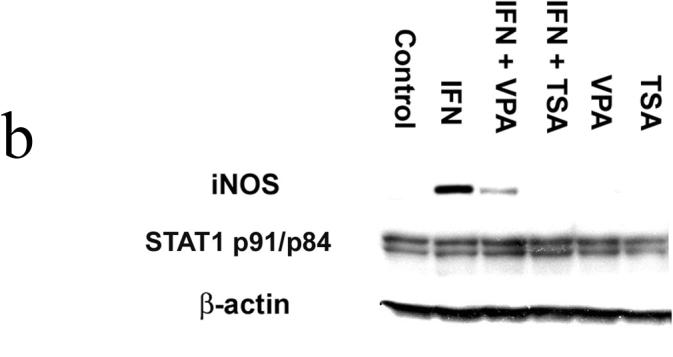
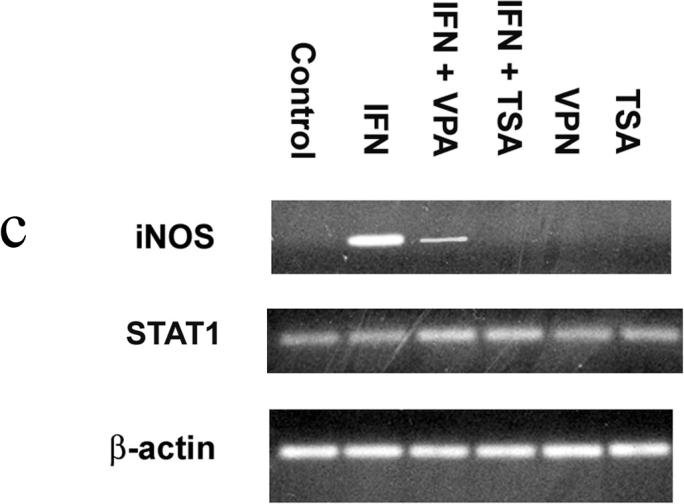
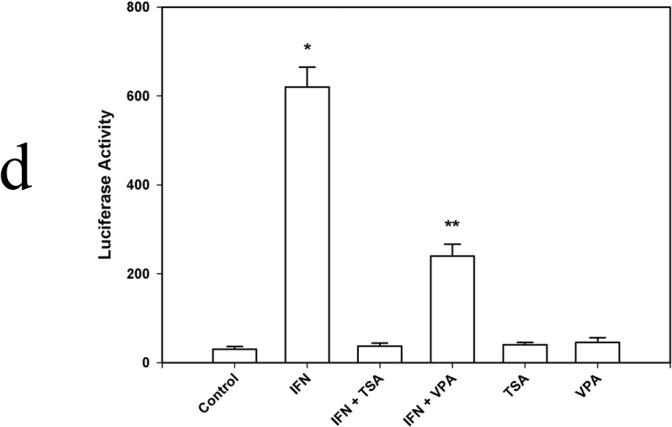
Figure 1a. NO synthesis. IFN-γ (500 u) was used to induce NO synthesis. In selected instances, the deacetylase inhibitors trichostatin A (TSA; 200 nM) and valproic acid (VPA; 1.5 mM) were used. After incubation for 3, 12, and/or 24 hr, 50 µl of culture medium was mixed with 50 µl of 1% sulfanilamide dissolved in 0.5 mol/L HCl. After 5-min incubation at room temperature, 50 µl of N-(1-naphthyl)-ethylenediamine was added. Following incubation for 10 min at room temperature, the absorbance of samples was measured at 540 nm and compared with NaNO3 standards. Data are presented as mean ± SEM of four separate experiments. (* p<0.01 IFN vs. IFN + TSA and IFN + VPA; ** p<0.01 IFN + VPA vs. IFN and VPA)
Figure 1b. Stat1 and iNOS protein expression. Cells were lysed, homogenized, and centrifuged. The cell lysates were separated by SDS-PAGE and transferred to PVDF membrane. Blocked membranes were then incubated with iNOS, β-actin , or Stat1 p84/p91 antibody. After washing, the membranes were incubated with donkey anti-rabbit IgG-horse radish peroxidase. Protein-antibody complexes were visualized using chemiluminescence techniques. Blot is representative of three experiments.
Figure 1c. Stat1 and iNOS mRNA. Total RNA was isolated from RAW 264.7 macrophages; mRNA (0.5 µg) were reverse-transcribed into cDNA. The primers for iNOS were 5′-CATCCATGCAAAGAACGTGT-3′ (forward) and 5′-GAAGGTGAGCTG AACGAGGA-3′ (reverse), and the primers for Stat1 were 5′-CTTATTCCATGGA CAAGGTTTTG-3′ (forward) and 5′-GGTGCTTCTTAATGAGCTCTAGG-3′ (reverse). The intensities of the PCR products were normalized to that of housekeeping gene β-actin. Gel is representative of three experiments.
Figure 1d. Transient transfection analysis of the iNOS promoter. 1×106 cells were allowed to grow for 24 h before the transfection. 2µg plasmid DNA and 2µg protamine sulfate diluted in OPTI-DMEM and 24 ug lipofectamine diluted in OPTI-DMEM were combined and incubated at room temperature for 20 min. To control transfection efficiency between groups, 0.1 ug pRL-TK was added to each well. Cells were harvested in 0.4 ml of reporter lysis buffer, and dual luciferase reporter assays were performed. Data are presented as mean ± SEM of four separate experiments. (* p<0.01 IFN vs. IFN + TSA and IFN + VPA; ** p<0.01 IFN + VPA vs. IFN and VPA)
As Stat1 is critical for IFN-γ mediated iNOS transcription, we next examined the effect of VPA and TSA upon Stat1 acetylation. Co-IP assays were performed using antibody to acetylated lysine moieties for IP, followed by immunoblotting for Stat1. (Figure 2) Minimal levels of acetylated Stat1 were identified in Controls. IFN-γ increased this level of expression by 5-fold. (p<0.01 vs. Control) The level of acetylated Stat1 was was further increased in IFN + TSA by an additional 4-fold. (p<0.02 vs. IFN) We then performed immunoblots for phosphorylated Stat1 under the same treatment conditions. Interestingly, addition of VPA or TSA to IFN treated cells was associated with progressively increased levels of activated (and phosphorylated Tyr 701) Stat1. Again, total Stat1 levels were not altered under any of our stimulation conditions. To address the potential for accelerated Stat1 degradation in the presence of TSA, we performed serial immunoblots in IFN-γ stimulated cells following addition of cycloheximide, an inhibitor of translation. In this setting, there was not alteration in Stat1 protein half-life in IFN vs IFN + TSA stimulated cells, 7.8 ± 1.2 hrs vs 8.4 ± 2.0 hrs, respectively.
Figure 2. Immunoprecipitation of Ac-Stat1 from IFN-γ stimulated macrophages.
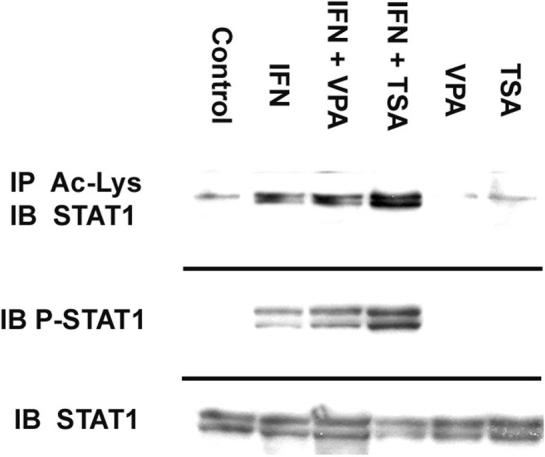
750 µg of extracted protein were mixed with 500 µl of PBS, 1 µl of normal mouse IgG and 20 µl of G-plus agarose. Anti-acetylated-lysine was added, followed by G-plus agarose. Protein was analyzed by western blotting with Stat1 p84/p91 antibody. In separate experiments, immunblots were performed for total Stat1 and phosphorylated Tyr701 Stat1 (P-Stat1). The cell lysates were separated by SDS-PAGE and transferred to PVDF membrane. Blocked membranes were then incubated with Stat1 p84/p91 or Tyr 701 Stat1 antibody. Blot is representative of three experiments.
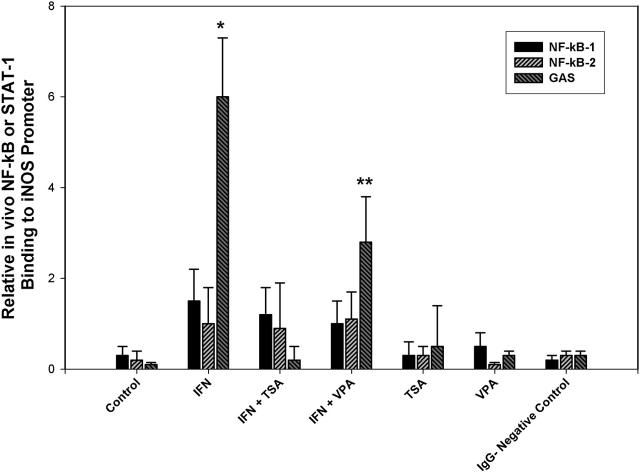
Finally, real time PCR-ChIP assays were performed to determine the effect of global deacetylase inhibition upon Stat1 binding to the iNOS promoter. (Figure 3) As has been previously demonstrated by Ganster et. al., NF-κB binding was minimal compared to that of Stat1 in the setting of IFN-γ stimulation.(7) Stat1 binding was readily detected in the presence of IFN. However, in IFN + TSA cells, Stat1 binding is decreased to levels equivalent to that of unstimulated Controls. (p<0.01 IFN + TSA vs IFN) Again, IFN + VPA also significantly decreased Stat1 binding in comparison to IFN, but to a lesser extent that that noted in IFN + TSA cells. (p<0.01 IFN + VPA vs IFN and IFN + TSA) These results suggest that Stat1 acetylation is associated with decreased Stat1 binding to the iNOS promoter despite the concomitant increased levels of phosphorylated Stat1. In addition, this points out the relatively minor role played by NF-κB in the initiation of iNOS transcription in IFN-γ stimulated macrophages.
Figure 3. Stat1 and NF-κB binding to the iNOS promoter.
Chromatin from macrophages was fixed and immunoprecipitated. Purified chromatin was immunoprecipitated using anti-NF-κB p65, anti-Stat1 or rabbit nonimmune serum; eluted DNA fragments were purified to serve as templates. The input fraction corresponded to 0.1 and 0.05% of the chromatin solution before immunoprecipitation. After DNA purification, the presence of the selected DNA sequence was quantified by real time-PCR. Data are presented as mean ± SEM of four separate experiments. (* p<0.01 IFN vs. IFN + TSA and IFN + VPA; ** p<0.01 IFN + VPA vs. IFN and VPA)
In association with our previous results examining iNOS protein, mRNA and promoter activation, these data indicate that Stat1 acetylation is associated with decreased Stat1 binding to the iNOS promoter and decreased iNOS expression. Acetylation of Stat1 may serve to down regulate macrophage iNOS expression in pro-inflammatory states.
Discussion
In the present study, we demonstrate that acetylation of Stat1 in IFN-γ stimulated RAW 264.7 murine macrophages accompanies decreased binding to the iNOS promoter GAS binding site, decreased iNOS mRNA and protein expression, and decreased NO production. This occurs despite increased levels of activated Stat1 that is phosphorylated at Tyr 701. Stat1 Tyr701 phosphorylation induces Stat1 dimerization, nuclear translocation and DNA binding. This suggests that a hierarchy of acetylation over phosphorylation may exist in this system.
The iNOS promoters of different species exhibit homologies to binding sites for numerous transcription factors (such as activating protein-1; CCAAT-enhancer box binding protein; cAMP-responsive element binding protein; interferon regulatory factor-1; NF-κB; nuclear factor-IL6; octamer factor-1; serum response factor; and Stat1) known to be involved in the LPS/cytokine-mediated induction of transcription.(8;9) All mammalian iNOS promoters contain several homologies with the IFN-γ-regulated transcription factor Stat1α binding sites (GAS). An essential role for the Stat1α pathway in iNOS induction had been shown for murine, rat and human cells. Stat proteins are well characterized as critical mediators of cytokine signaling. Generally, the Stat signaling pathway is triggered by binding of cytokines to specific cell surface receptors.(10;11) JAK-activity phosphorylates tyrosine residues in the cytoplasmic portions of the cytokine receptor, which in turn serves as a docking site for the latent Stat proteins. Thereafter, Stat proteins are also phosphorylated, translocate as dimers into the nucleus, bind to target DNA sequences of promoter regions, and transactivate gene transcription. Thereafter, the dephosphorylated Stat protein returns to the cytoplasm. Several compounds have been described that inhibit iNOS expression by blocking cytokine-induced activation of Stat1α.(12-14) Gao reported that binding of Stat1α to the GAS of the murine iNOS promoter was required for optimal induction of the iNOS gene by IFN-γ and LPS.(15) Also, the IFN-γ-mediated enhancement of IL-1β-induced promoter activity in rat RINm5F cells or rat aortic smooth muscle cells was dependent on the GAS and ISRE sites around position −900 bp of the rat iNOS promoter.(16;17) In human A549 or DLD1 cells, inhibition of the IFN-γ-activated tyrosine kinase JAK2 by tyrphostin B42 (AG 490) reduced Stat1α DNA-binding activity and iNOS expression.(18) Using specific signaling inhibitors, Blanchette et al. showed that JAK2/Stat1α- and extracellular regulated kinase-(ERK1/ERK2)-dependent pathways are critical components of IFN-γ-inducible iNOS expression in murine J774 macrophages.(19) A dominant-negative mutant of Stat1α inhibited cytokine-induced human iNOS promoter activity.(20) Site-directed mutagenesis of the human iNOS promoter identified a bifunctional NF-κB/Stat1α motif at −5.8 kb, and a Stat1α-specific responsive element at −5.2 kb.(20) iNOS induction is blocked in macrophages from mice with a disrupted Stat1α gene.(21) Thus, the bulk of experimental data indicates that Stat1 is an essential activator of IFN-γ stimulated iNOS transcription.
The link between histone acetylation and regulation of transcription has been known for many years. However, recently it has become clear that a number of non-histone proteins serve as targets for nuclear histone acetylases, including p53, GATA1 and α-tubulin.(6) Substrates include such as DNA binding proteins, non-buclear proteins and proteins that shuttle from the nucleus to the cytoplasm. As a consequence, acetylation may alter DNA binding specificity of transcription factors depending upon the the relative location of the acetylation site to the DNA binding domain. Acetylation may therefore stimulate or inhibit transcription. In addition, acetylation may regulate protein-protein interactions or protein stability. Overall, the relationship between acetylation and phosphorylation has not been extensively characterized and represents the complexity of overlapping signalling pathways. In the case of PC4, phosphorylation overrides acetylation with regard to activation.(22)
While IFN-γ certainly induces Stat1 activation/phosphorylation, the acetylation of Stat1 may also occur as a counter regulatory mechanism to ultimately aid in “turning off” iNOS transcription. Inhibition of deacetylase activity by TSA or to a lesser extent, VPA, are associated with decreased IFN mediated iNOS protein, mRNA and promoter activity. This occurs in the setting of increased Stat1 acetylation and despite increased Stat1 phosphorylation, suggesting a hierarchy of post-translational modification. Finally, in our real time ChIP assays, there is decreased in vivo binding of Stat1 to the GAS element of the iNOS promoter in the presence of increase Ac-Stat1. Again, this occurs despite increased Stat1 phosphorylation and maintenance of total cellular Stat1.
Acknowledgments
Work supported by NIH grants GM65113 (PCK), AI44629 (PCK) and DK070642 (PCK).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Xie QW, Nathan C. The high output nitric oxide pathway: role and regulation. Journal of Leukocyte Biology. 1994;56:576–82. doi: 10.1002/jlb.56.5.576. [DOI] [PubMed] [Google Scholar]
- 2.Doursout MF, Kilbourn RG, Hartley CJ, Chelly JE. Effects of N-methyl-L-arginine on cardiac and regional blood flow in a dog endotoxin shock model. J Crit Care. 2000;15:22–9. doi: 10.1053/jcrc.2000.0150022. [DOI] [PubMed] [Google Scholar]
- 3.Kilbourn RG, Traber DL, Szabo C. Nitric oxide and shock. Dis Mon. 1997;43:277–348. doi: 10.1016/s0011-5029(97)90028-6. [DOI] [PubMed] [Google Scholar]
- 4.Oddis CV, Simmons RL, Hattler BG, Finkel MS. Chronotropic effects of cytokines and the nitric oxide synthase inhibitor, L-NMMA, on cardiac myocytes. Biochem Biophys Res Commun. 1994;205:992–7. doi: 10.1006/bbrc.1994.2764. [DOI] [PubMed] [Google Scholar]
- 5.Schindler C, Darnell JE., Jr. Transcriptional responses to polypeptide ligands: the JAKSTAT pathway. Annu Rev Biochem. 1995;64:621–51. doi: 10.1146/annurev.bi.64.070195.003201. [DOI] [PubMed] [Google Scholar]
- 6.Kouzarides T. Acetylation: a regulatory modification to rival phosphorylation? EMBO J. 2000;19:1176–9. doi: 10.1093/emboj/19.6.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ganster RW, Guo Z, Shao L, Geller DA. Differential effects of TNF-alpha and IFN-gamma on gene transcription mediated by NF-kappaB-Stat1 interactions. J Interferon Cytokine Res. 2005;25:707–19. doi: 10.1089/jir.2005.25.707. [DOI] [PubMed] [Google Scholar]
- 8.Lin AW, Chang CC, McCormick CC. Molecular cloning and expression of an avian macrophage nitric-oxide synthase cDNA and the analysis of the genomic 5'-flanking region. J Biol Chem. 1996;271:11911–9. doi: 10.1074/jbc.271.20.11911. [DOI] [PubMed] [Google Scholar]
- 9.Chu SC, Marks-Konczalik J, Wu HP, Banks TC, Moss J. Analysis of the cytokine-stimulated human inducible nitric oxide synthase (iNOS) gene: characterization of differences between human and mouse iNOS promoters. Biochem Biophys Res Commun. 1998;248:871–8. doi: 10.1006/bbrc.1998.9062. [DOI] [PubMed] [Google Scholar]
- 10.Shuai K, Liu B. Regulation of JAK-STAT signalling in the immune system. Nat Rev Immunol. 2003;3:900–11. doi: 10.1038/nri1226. [DOI] [PubMed] [Google Scholar]
- 11.Platanias LC. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat Rev Immunol. 2005;5:375–86. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- 12.Tedeschi E, Menegazzi M, Margotto D, Suzuki H, Forstermann U, Kleinert H. Anti-inflammatory actions of St. John's wort: inhibition of human inducible nitric-oxide synthase expression by down-regulating signal transducer and activator of transcription-1alpha (STAT-1alpha) activation. J Pharmacol Exp Ther. 2003;307:254–61. doi: 10.1124/jpet.103.054460. [DOI] [PubMed] [Google Scholar]
- 13.Tedeschi E, Menegazzi M, Yao Y, Suzuki H, Forstermann U, Kleinert H. Green tea inhibits human inducible nitric-oxide synthase expression by down-regulating signal transducer and activator of transcription-1alpha activation. Mol Pharmacol. 2004;65:111–20. doi: 10.1124/mol.65.1.111. [DOI] [PubMed] [Google Scholar]
- 14.Yao Y, Hausding M, Erkel G, Anke T, Forstermann U, Kleinert H. Sporogen, S14−95, and S-curvularin, three inhibitors of human inducible nitric-oxide synthase expression isolated from fungi. Mol Pharmacol. 2003;63:383–91. doi: 10.1124/mol.63.2.383. [DOI] [PubMed] [Google Scholar]
- 15.Gao J, Morrison DC, Parmely TJ, Russell SW, Murphy WJ. An interferon-gamma-activated site (GAS) is necessary for full expression of the mouse iNOS gene in response to interferon-gamma and lipopolysaccharide. J Biol Chem. 1997;272:1226–30. doi: 10.1074/jbc.272.2.1226. [DOI] [PubMed] [Google Scholar]
- 16.Darville MI, Eizirik DL. Regulation by cytokines of the inducible nitric oxide synthase promoter in insulin-producing cells. Diabetologia. 1998;41:1101–8. doi: 10.1007/s001250051036. [DOI] [PubMed] [Google Scholar]
- 17.Teng X, Zhang H, Snead C, Catravas JD. Molecular mechanisms of iNOS induction by IL-1 beta and IFN-gamma in rat aortic smooth muscle cells. Am J Physiol Cell Physiol. 2002;282:C144–C152. doi: 10.1152/ajpcell.2002.282.1.C144. [DOI] [PubMed] [Google Scholar]
- 18.Kleinert H, Wallerath T, Fritz G, Ihrig-Biedert I, Rodriguez-Pascual F, Geller DA, et al. Cytokine induction of NO synthase II in human DLD-1 cells: roles of the JAK-STAT, AP-1 and NF-kappaB-signaling pathways. Br J Pharmacol. 1998;125:193–201. doi: 10.1038/sj.bjp.0702039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blanchette J, Jaramillo M, Olivier M. Signalling events involved in interferon-gamma-inducible macrophage nitric oxide generation. Immunology. 2003;108:513–22. doi: 10.1046/j.1365-2567.2003.01620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ganster RW, Taylor BS, Shao L, Geller DA. Complex regulation of human inducible nitric oxide synthase gene transcription by Stat 1 and NF-kappa B. Proc Natl Acad Sci U S A. 2001;98:8638–43. doi: 10.1073/pnas.151239498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meraz MA, White JM, Sheehan KC, Bach EA, Rodig SJ, Dighe AS, et al. Targeted disruption of the Stat1 gene in mice reveals unexpected physiologic specificity in the JAK-STAT signaling pathway. Cell. 1996;84:431–42. doi: 10.1016/s0092-8674(00)81288-x. [DOI] [PubMed] [Google Scholar]
- 22.Kumar BRP, Swaminathan V, Banerjee S, Kundu TK. P300-mediated acetylation of human transcriptional coactivator PC4 is inhibited by phosphrylation. J Biol Chem. 2001;276:16804–16809. doi: 10.1074/jbc.M100934200. [DOI] [PubMed] [Google Scholar]