Figure 1. Acetylation and NO synthesis in IFN-γ stimulated RAW264.7 murine macrophages.
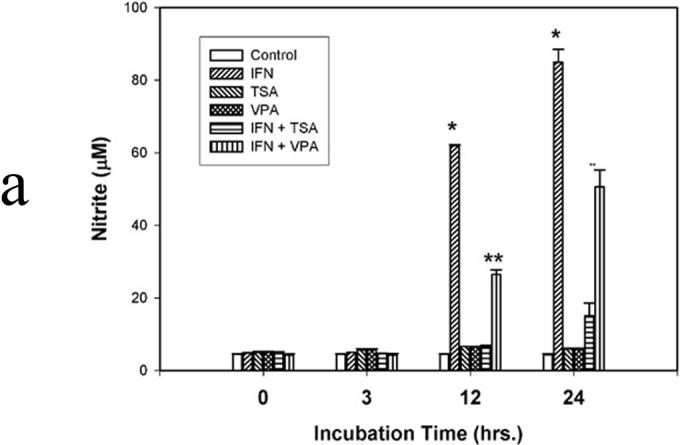
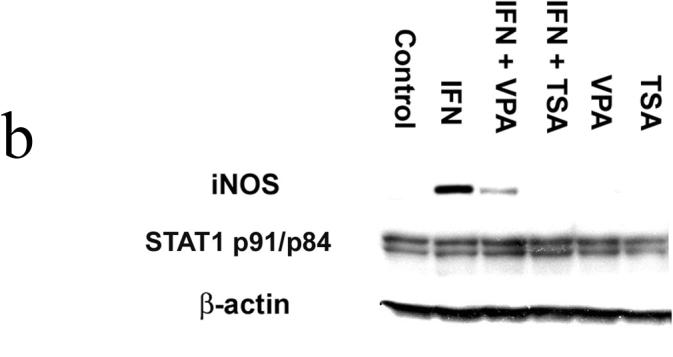
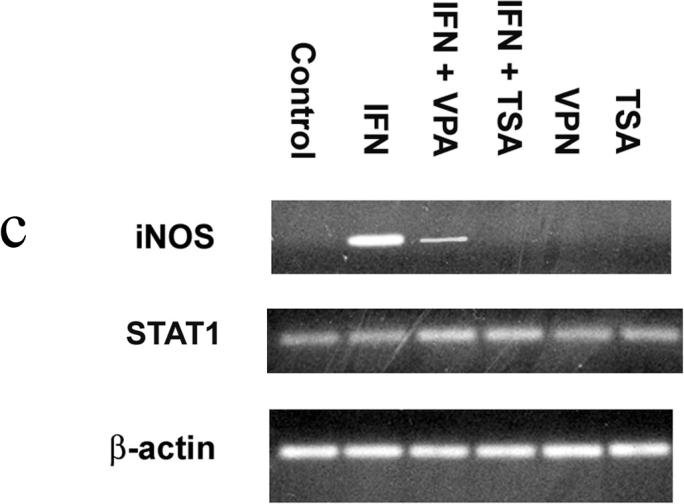
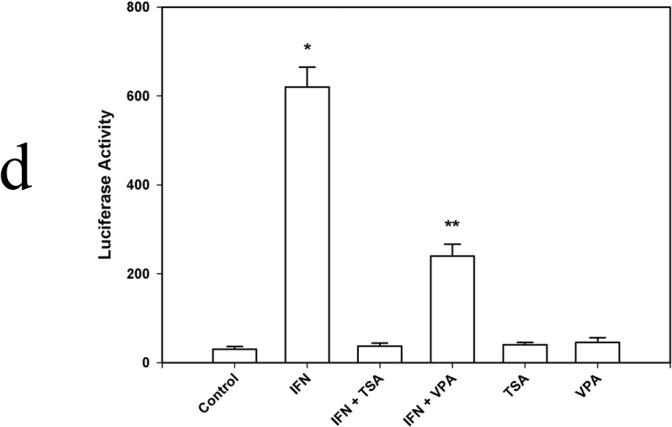
Figure 1a. NO synthesis. IFN-γ (500 u) was used to induce NO synthesis. In selected instances, the deacetylase inhibitors trichostatin A (TSA; 200 nM) and valproic acid (VPA; 1.5 mM) were used. After incubation for 3, 12, and/or 24 hr, 50 µl of culture medium was mixed with 50 µl of 1% sulfanilamide dissolved in 0.5 mol/L HCl. After 5-min incubation at room temperature, 50 µl of N-(1-naphthyl)-ethylenediamine was added. Following incubation for 10 min at room temperature, the absorbance of samples was measured at 540 nm and compared with NaNO3 standards. Data are presented as mean ± SEM of four separate experiments. (* p<0.01 IFN vs. IFN + TSA and IFN + VPA; ** p<0.01 IFN + VPA vs. IFN and VPA)
Figure 1b. Stat1 and iNOS protein expression. Cells were lysed, homogenized, and centrifuged. The cell lysates were separated by SDS-PAGE and transferred to PVDF membrane. Blocked membranes were then incubated with iNOS, β-actin , or Stat1 p84/p91 antibody. After washing, the membranes were incubated with donkey anti-rabbit IgG-horse radish peroxidase. Protein-antibody complexes were visualized using chemiluminescence techniques. Blot is representative of three experiments.
Figure 1c. Stat1 and iNOS mRNA. Total RNA was isolated from RAW 264.7 macrophages; mRNA (0.5 µg) were reverse-transcribed into cDNA. The primers for iNOS were 5′-CATCCATGCAAAGAACGTGT-3′ (forward) and 5′-GAAGGTGAGCTG AACGAGGA-3′ (reverse), and the primers for Stat1 were 5′-CTTATTCCATGGA CAAGGTTTTG-3′ (forward) and 5′-GGTGCTTCTTAATGAGCTCTAGG-3′ (reverse). The intensities of the PCR products were normalized to that of housekeeping gene β-actin. Gel is representative of three experiments.
Figure 1d. Transient transfection analysis of the iNOS promoter. 1×106 cells were allowed to grow for 24 h before the transfection. 2µg plasmid DNA and 2µg protamine sulfate diluted in OPTI-DMEM and 24 ug lipofectamine diluted in OPTI-DMEM were combined and incubated at room temperature for 20 min. To control transfection efficiency between groups, 0.1 ug pRL-TK was added to each well. Cells were harvested in 0.4 ml of reporter lysis buffer, and dual luciferase reporter assays were performed. Data are presented as mean ± SEM of four separate experiments. (* p<0.01 IFN vs. IFN + TSA and IFN + VPA; ** p<0.01 IFN + VPA vs. IFN and VPA)
