Figure 3.
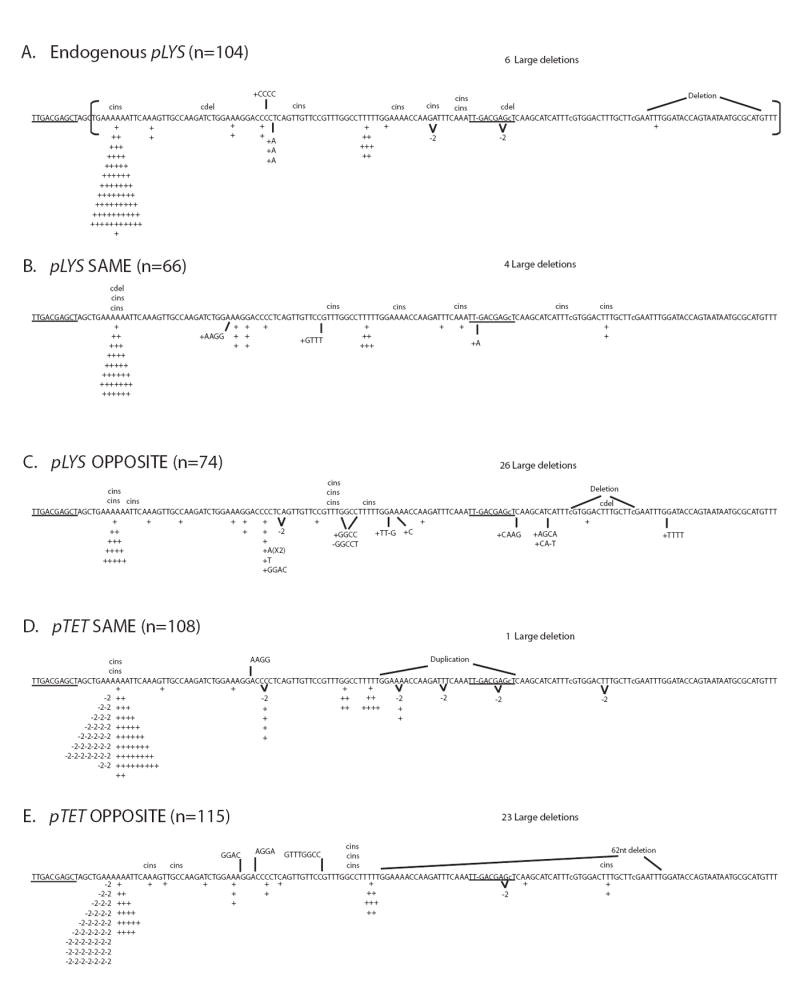
Reversion spectra for lys2ΔA746 allele under low-transcription conditions (no Dox for pLYS alleles and 1000 ng/ml Dox for pTET alleles).
The sequence shown includes the reversion window and the 14 nts upstream (the reversion window begins with 5’TGAAAAAA… and is marked by a bracket); the position of the A nucleotide deleted to create the lys2ΔA746 allele is indicated by a dash and the nucleotides changed to extend the reversion window are indicated by lowercase letters [42]. 1-nt insertions and 2-nt deletions are indicated under the sequence by “+” or “-2”, respectively. Complex insertions (cins) and deletions (cdel) are indicated above the sequence. The number of 95-nt deletion events are indicated as “large dels” above each spectrum. The 10-nt direct repeat sequences at the end points of the large dels are underlined. An abbreviated designation for each allele is used; pLYS same is pLYS-lys2ΔA746 in the SAME orientation, etc. The total number of revertants sequenced (n) for each strain is indicated. The reversion spectrum for pLYS-lys2ΔA746 allele at its endogenous location was previously published [24].
