TABLE 2.
Medians (5th, 95th percentiles) of the estimates of mutation rate, migration rate, and effective population size using genotype data simulated under the finite-island model with the k-allele mutation model at different values of mutation rate, u, migration rate, m, and effective population size, N, based on 400 replicate simulations for each pair of model migration rate and effective population size parameters for dioecious (u = 0.001, u = 0.0005, and u = 0.0001) and monoecious (u = 0.0005) model populations
| Parameter values
|
Parameter estimates
|
|||
|---|---|---|---|---|
| N | m |
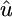 × 103 × 103
|
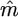 |
 |
| Dioecious populations: k-allele mutation model, u = 0.001 | ||||
| 10 | 0.02 | 0.993 (0.693, 1.343) | 0.019 (0.013, 0.027) | 9.97 (8.36, 12.53) |
| 0.20 | 1.003 (0.716, 1.425) | 0.201 (0.155, 0.260) | 10.02 (8.60, 11.80) | |
| 20 | 0.02 | 1.005 (0.675, 1.348) | 0.020 (0.014, 0.026) | 20.09 (15.83, 26.87) |
| 0.20 | 1.005 (0.732, 1.371) | 0.202 (0.152, 0.257) | 19.92 (16.72, 24.95) | |
| 50 | 0.02 | 0.989 (0.523, 1.507) | 0.019 (0.011, 0.029) | 50.83 (35.17, 88.58) |
| 0.20 | 0.971 (0.569, 1.524) | 0.196 (0.112, 0.305) | 50.82 (35.10, 83.54) | |
| Parameter values
|
Parameter estimates
|
|||
| N | m |
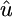 × 104 × 104
|
 |
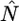 |
| Dioecious populations: k-allele mutation model, u = 0.0005 | ||||
| 10 | 0.02 | 4.937 (3.299, 7.462) | 0.019 (0.014, 0.028) | 10.09 (8.12, 12.76) |
| 0.20 | 5.100 (3.287, 7.125) | 0.200 (0.144, 0.270) | 9.89 (8.50, 12.52) | |
| 20 | 0.02 | 5.038 (3.355, 7.123) | 0.020 (0.014, 0.026) | 19.97 (15.30, 27.83) |
| 0.20 | 4.914 (3.413, 7.067) | 0.198 (0.145, 0.265) | 19.86 (16.50, 25.72) | |
| 50 | 0.02 | 5.018 (2.529, 8.093) | 0.020 (0.010, 0.030) | 49.36 (33.45, 98.02) |
| 0.20 | 5.034 (2.521, 7.748) | 0.199 (0.098, 0.314) | 49.98 (34.53, 92.92) | |
| Dioecious populations: k-allele mutation model, u = 0.0001 | ||||
| 10 | 0.02 | 0.970 (0.423, 1.677) | 0.019 (0.010, 0.028) | 9.94 (7.59, 16.63) |
| 0.20 | 0.962 (0.354, 1.734) | 0.200 (0.105, 0.327) | 9.97 (7.47, 15.78) | |
| 20 | 0.02 | 0.977 (0.444, 1.610) | 0.019 (0.010, 0.030) | 19.68 (13.78, 35.61) |
| 0.20 | 1.025 (0.446, 1.824) | 0.198 (0.106, 0.339) | 19.92 (13.19, 35.27) | |
| 50 | 0.02 | 0.919 (0.096, 1.809) | 0.018 (0.002, 0.034) | 53.17 (29.94, 447.1) |
| 0.20 | 0.933 (0.250, 1.736) | 0.191 (0.056, 0.374) | 51.81 (31.04, 172.7) | |
| Monoecious populations: k-allele mutation model, u = 0.0005 | ||||
| 10 | 0.02 | 4.965 (3.299, 6.878) | 0.019 (0.014, 0.026) | 10.07 (8.69, 12.34) |
| 0.20 | 5.046 (3.356, 7.129) | 0.198 (0.151, 0.260) | 10.08 (8.77, 11.84) | |
| 20 | 0.02 | 4.985 (3.266, 7.065) | 0.020 (0.013, 0.027) | 19.83 (15.65, 27.23) |
| 0.20 | 4.836 (3.499, 7.004) | 0.196 (0.143, 0.266) | 20.17 (16.13, 26.19) | |
| 50 | 0.02 | 5.108 (2.216, 7.818) | 0.020 (0.009, 0.030) | 49.68 (33.53, 110.1) |
| 0.20 | 4.863 (2.877, 7.566) | 0.201 (0.113, 0.309) | 50.55 (35.33, 84.27) | |
