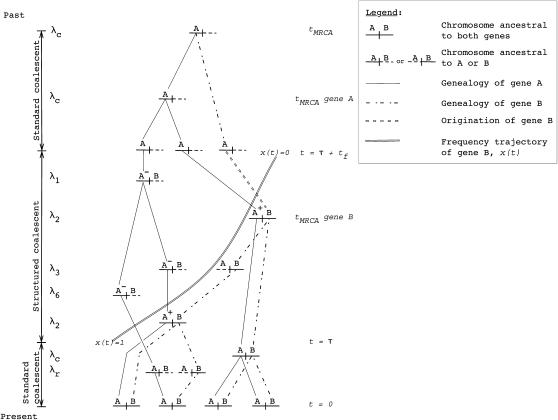
Figure 3.—
Example of a gene genealogy for partially linked, duplicated genes. A sample of size n = 4 is followed back to the most recent common ancestor (MRCA) of both genes. Gene B, the recent duplicate, fixed at time τ in the past, and an “A” label represents the ancestral gene. Prior to τ, the genealogical process is the standard coalescent for two partially linked loci. At time τ, the simulation enters a structured coalescent phase, during which there are two types of chromosomes in the history of gene A. First, at any time t during the structured phase, there are chromosomes whose ancestry is in the part of the population ancestral to the duplicate. These are labeled A+. The second type has an ancestry in the portion of the population not containing the duplicate and is labeled A−. Crossing over between loci can move chromosomes between these two classes (see simulation). Note that the A+ and A− labels are necessary only during the structured phase, where one must keep track of rates of coalescence within subpopulations of different sizes. The MRCA of B is guaranteed to be reached during the structured phase, and the MRCA of B is then considered to be an allele of gene A, i.e., the mutation event that gave rise to B. After the structured phase, any remaining lineages are followed back to their MRCA according to the standard coalescent process. To the left of the recombination graph are the rates that gave rise to the chromosomes shown on the genealogy. The rates correspond to Equations 6–17.