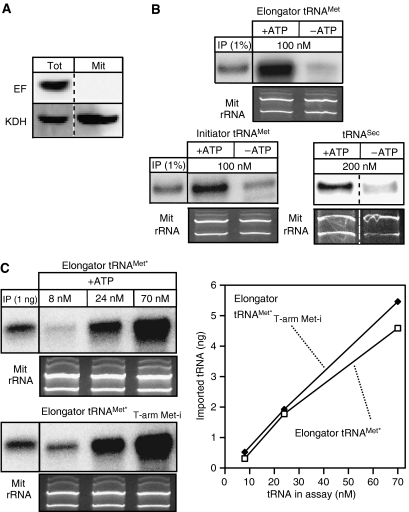
Figure 6.
In vitro import assays. (A) Immunoblot analysis of 25 mg each of total cellular extract (Tot) and purified mitochondria (Mit) used in the in vitro assays for the presence of eEF1a (EF, top panel) and the mitochondrial marker α-ketoglutarate dehydrogenase (KDH, bottom panel), respectively (B) Left and middle panels: In vitro import in the presence and absence of ATP of in vitro transcribed and radioactively labeled imported elongator tRNAMet or cytosol-specific initiator tRNAMet, respectively. The input lanes depict 1% of the added substrate. Right panel: In vitro import of in vitro transcribed cytosol-specific tRNASec. In this case the imported tRNASec was detected by Northern blot and specific oligonucleotide hybridization. The concentration of each substrate tRNA in the import reaction is indicated. (C) In vitro import assays containing ATP and the indicated concentrations of in vitro transcribed tagged in vivo in part imported elongator tRNAMet (elongator tRNAMet*, top panel) or an in vivo cytosol-specific variant thereof carrying the T-arm of the initiator tRNAMet (elongator tRNAMet*T−arm Met−i, bottom panel). Imported tRNAs were detected by Northern blots and hybridization of oligonucleotides directed against the tag (Crausaz-Esseiva et al, 2004a). The input lanes show the hybridization signals obtained by 1 ng of substrate. The graph shows the quantification of the Northern blots. All import reactions shown in panels B and C were treated with micrococcus nuclease. Ethidium bromide-stained panels show the two mitochondrial rRNAs and serve as loading controls.