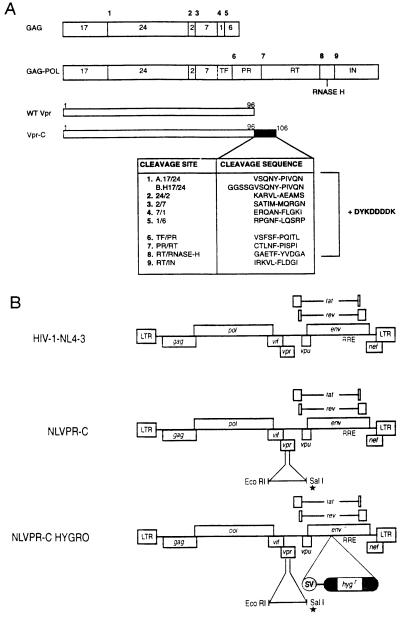
Figure 2.
(A) Schematic representation of the Gag and Gag–Pol precursors of HIV-1 indicating protease cleavage sites 1–9 (bold). The Vpr-C proteins contain the corresponding cleavage signal found in the Gag and Gag–Pol precursors indicated by number and abbreviation for the site, fused in-frame to the C terminus of Vpr resulting in a protein of 106 aa. Likewise, all Vpr-C constructs received a Flag epitope (DYKDDDDK) immediately following the cleavage site. The asterisk indicates the construct containing the Flag epitope alone. The dashed line indicates site of ribosomal frame shift. (B) Proviral clone pNL4–3 cleaved with EcoRI/SalI in Vpr coding region to allow insertion of EcoRI/XhoI generated fragment from Vpr-C (NLVPR-C). Vpr-C-derived fragments contain a stop codon (∗) so sequences downstream of SalI are out of frame. NLVPR-C HYGRO contains an SV-Hygr cassette in the env gene for selection of positive clones in single-round replication assay.