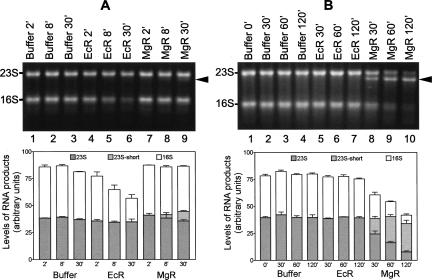
FIGURE 4.
Degradation of E. coli rRNA by MgR and EcR. Isolated rRNA was treated with RNases in Zn2+-containing buffer for the length of time indicated on the top of each lane. Agarose gels (1.2%) were used to separate the RNA. A photograph was taken under UV lights after staining with SYBR Gold. Intermediate 23S RNA degradation products are labeled on the right side by arrows. The RNA products were quantified using the integrated optical density of the bands detected by the Epi Chemi II Darkroom (UVP Laboratory Products). Gels from three independent runs were quantified, and average values with standard errors are shown in the bottom panels. (A) Reactions with equal amounts of MgR and EcR (0.15 μg each). (B) Reactions with MgR and EcR of similar poly(A) degradation activity (0.5 μg of MgR or 0.038 μg of EcR).