Abstract
We present here the use of fluorescent methodologies for structural and functional studies of RNA in place of radioactivity. The methods are highly sensitive and quantitative with the use of an infrared fluorescence imaging system. IRD-700 and IRD-800 labels are used for fluorescence detection. Chemical probing methods are largely used for mapping RNA secondary structure and to monitor ligand interactions and conformational changes involving individual bases of RNA. The new fluorescent primer extension methodology allows simple and fast chemical probing of RNA with high sensitivity. IRD-700 and IRD-800 labeled primers can also be used to monitor protein–RNA interactions by fluorescent mobility shift assays. The speed and ease of these approaches are advantages over prior methods that used hazardous radioisotopes. Structural and biochemical investigations of RNA should benefit from the use of these fluorescent methodologies.
Keywords: chemical probing, RNA–protein interaction, RNA–ligand interaction, reverse transcription, SHAPE, fluorescent primers
INTRODUCTION
Traditional chemical and enzymatic reagents and backbone-based cleavage are useful for investigating RNA secondary and tertiary structures (Peattie and Gilbert 1980; Ehresmann et al. 1987; Stern et al. 1988; Soukup and Breaker 1999; Hanson et al. 2005; Merino et al. 2005). These methods also provide insights into RNA–ligand interactions as well as conformational changes at single nucleotide resolution. These techniques require reverse transcriptase to extend either 32P- or 33P-labeled primers or incorporate radioactive NTPs internally. Otherwise, direct labeling of the RNA is required. While these approaches have been successful, they have the disadvantage of using hazardous radioisotope molecules and requiring time-consuming autoradiographical treatments. Substitution of radioactivity by fluorescent methodologies with similar sensitivity would be a significant improvement. We report here the fluorescent primer extension (FPE) methodology for RNA biochemical studies that takes advantage of the use of fluorescently labeled primers. We use DNA primers labeled with infrared dyes (IRD-700 or IRD-800). These dyes are used for automated DNA sequencers because of the high sensitivity due to very low background at infrared wavelengths (Middendorf et al. 1992; Williams and Soper 1995; Machida et al. 1997). Near-infrared detection is more sensitive than visible fluorescence. At longer wavelengths, biomolecules exhibit greatly reduced autofluorescence, resulting in lower background and enhanced sensitivity when infrared fluorophores are used for detection. These dyes have excellent water solubility and minimal nonspecific binding (Li-COR Biosciences). They also have the advantage of being stable for a long time, whereas radioactive 33P or 32P labels have a short half-life. The FPE methodology can be used to sequence RNA molecules, measure the extent of chemical modification within a modified RNA, and monitor ligand–RNA interactions. We also show that dissociation constants of protein–RNA complexes can be rapidly measured by hybridization of fluorescently labeled primers to the RNA molecules of interest, which allows visualization of the free and bound fraction of RNA in a mobility shift assay. The experiments use common polyacrylamide gel electrophoresis for the analysis. Labeling with fluorescent markers allows direct optical scanning of the gel after the electrophoresis and does not require fixation, drying, and autoradiographical treatments. These faster and safer approaches should greatly aid biochemical and structural investigations of RNA and RNA–ligand interactions.
RESULTS AND DISCUSSION
Fluorescent primer extension (FPE) of RNA
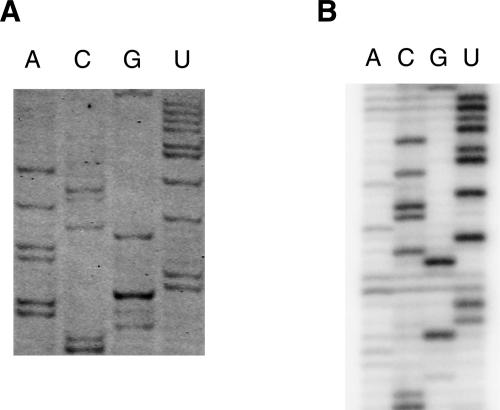
Primer extension using reverse transcriptase is a method used to analyze sequences of RNAs, to determine structural elements within RNA after chemical or enzymatic probing, and to investigate protein–RNA or ligand–RNA interactions by footprinting. To test the FPE method, we used Escherichia coli or yeast ribosomal RNAs as templates and performed direct sequencing reactions. IRD-700 and IRD-800 fluorescent DNA primers were purchased from MWG biotech. They are also available from other suppliers such as Li-COR Biosciences, Biolego, Thermo Hybaid, Metabion, and VH Bio. Alternatively, IRDye phosphoramidites are available from Li-COR Biosciences, allowing one to produce infrared-labeled primers on a DNA synthesizer in-house. For IRD-700, the extinction coefficient is 170,000 M−1 cm−1 with maximal absorption at 685 nm and maximal emission at 705 nm. For IRD-800 the extinction coefficient is 200,000 M−1 cm−1 with maximal absorption at 787 nm and maximal emission at 807 nm. At the moment and to the best of our knowledge, these fluorescent labels cannot be added enzymatically but need to be incorporated chemically during oligonucleotide synthesis. An example of an FPE experiment in which we used a DNA primer labeled at its 5′ end with IRD-700 and complementary to bases 1530–1509 of 16S rRNA is shown in Figure 1A. For comparison, a conventional primer extension of the same sequence using the same primer labeled with 32P radioisotope is shown in Figure 1B. The same quantity of the RNA template (0.3 μg of 16S rRNA) and the primers (1 μL of 10 μM primer) were used. The products of the primer-extension reactions were separated on 8% acrylamide–7 M urea gel in identical conditions of migration. For FPE, the gel is kept away from direct exposure to light by using a simple cover. Similar sequencing patterns were obtained from the two different methods (Fig. 1).
FIGURE 1.
Sequencing of E. coli rRNA using an IRD700-labeled primer (A) or a 32P-labeled primer (B). (A) Fluorescent DNA molecules were observed with an infrared imaging system Odyssey (LI-COR Biosciences). (B) Radioactive bands were visualized using a PhosphorImager Storm (Molecular Dynamics).
It should be noted that one of the advantages of the FPE method is the speed and the relative ease of use. For the FPE experiment, the gel was scanned by an infrared imaging system (Odyssey, LI-COR), directly after electrophoresis. For the radioactive experiment, the gel was transferred to a piece of 3MM filter paper and dried under vacuum for 1 h. This step was then followed by typically a few hours to overnight exposure to the PhosphorImager screen before scanning the screen by an imaging system (Storm, GE Healthcare).
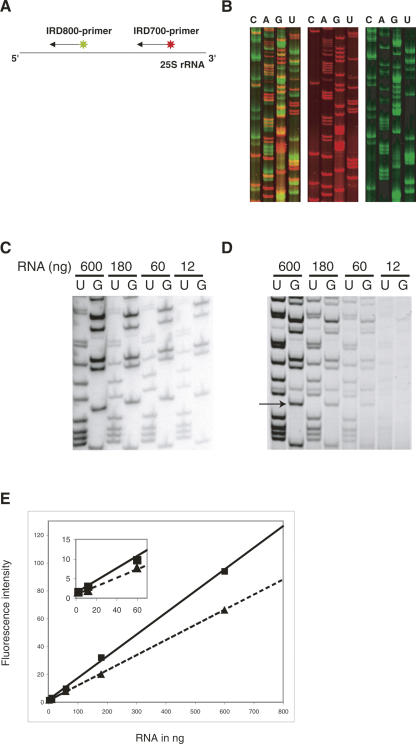
Furthermore, we tested the use of two primers that are labeled with two different fluorescent labels (IRD-700 and IRD-800) in a single FPE reaction. Several combinations of primers specific for various parts of rRNA were tested. As illustrated in Figure 2A, in a single reaction tube, two primers with different fluorescent labels were annealed to the same template (25S rRNA) and extended simultaneously. A part of the gel is shown in Figure 2B. Each primer resulted in a sequence of its corresponding area, in which red represents the sequence from nucleotides 388 to 439 and green from 92 to 139 of 25S rRNA. Compared with the reaction performed using a single primer, no dissimilarity was observed on the detection sensitivity and on the sequencing accuracy, although the detection of the IRD-700-labeled products (red) is much more efficient than the one for IRD-800-labeled cDNAs (Fig. 2B). The distance between two primers (Fig. 2A) was ∼300 nucleotides (nt). When the spacing between the two primers is shortened to ∼180 nt, the FPE reaction of the downstream primer is affected by the existence of the other primer and could not reach its longest extension (data not shown). On the other hand, the extension of the upstream primer is unaffected by the downstream one (data not shown). This suggests that attention must be paid to the interval between the two primers, and we recommend it to be longer than 300 nt. Another approach was carried out using two primers that target for two different rRNAs, 5.8S rRNA and 25S rRNA (data not shown). As expected, the FPE of both rRNAs was precisely achieved. The double-extension strategy can be advantageous when the quantity of the template is limited.
FIGURE 2.
Sequencing of yeast ribosomal RNAs using FPE. (A) Schematic drawing of the double extension strategy. (B) Fluorescent imaging of sequencing of rRNAs using FPE. (Left) The gel scanned at both 700 nm (red) and 800 nm (green) wavelengths; (middle) the results at 700 nm (red), and right, at 800 nm (green). (C) Sequencing reactions performed within a 50-fold dilution range from 600 to 12 ng of RNA using internal radioactive labeling of the cDNA. (D) The same experiment using a primer labeled with a single IRD-700. (E) Linearity of the fluorescence signal within a 250-fold dilution range of 600–2.4 ng of RNA. Dashed and solid lines correspond to two different bands of cDNA that were quantified. One of the bands that has been quantified is indicated by an arrow in D. (Inset) Enlargement of the crowded region from 0 to 70 ng.
cDNA extension products can also be separated by capillary electrophoresis or by slab gel electrophoresis using a DNA Analysis System (Beckman Coulter CEQ 8800XL) or 4300 DNA Analysis System (Li-COR Biosciences), respectively. Both systems present the advantage of directly obtaining a trace of the separated cDNAs (capillary electrophoresis) or an image of the gel (slab gel electrophoresis) without manipulation and scanning of the gel that is required with the Odyssey system.
Comparison of radioactivity and near-infrared fluorescence methods
In order to investigate the sensitivity of the infrared imaging system Odyssey, dilutions of an IRD-800-labeled primer were loaded on a native polyacrylamide gel. A band containing 0.5 fmol of primer could be easily detected. IRD-labeled DNA oligonucleotide detection is linear within a range of 10–0.2 fmol. We then performed primer extension experiments using radioactivity or FPE methods to directly address the sensitivity of both methods. In Figure 2C, sequencing reactions were performed within a 50-fold dilution range of 600–12 ng of RNA using internal radioactive labeling of the synthesized cDNA. The same experiment using primers labeled with single IRD-700 dyes is shown in Figure 2D. The intensity of two different cDNA bands was quantified for each RNA quantity tested. The IRD-labeled cDNA detection is linear within a 50-fold dilution range of 600–12 ng of RNA (Fig. 2E, and inset). The fluorescent method using a single fluorophore approached the sensitivity obtained with internal radioactive labeling that incorporates several molecules of a radioactive nucleotide within a cDNA. At the moment, the detection limit with fluorescence remains a factor of 10 above the limit reached with radioactivity.
RNA–ligand interactions monitored by chemical probing
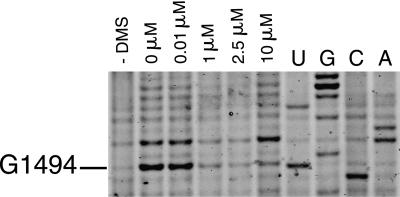
Chemical probing methods are largely used for mapping RNA secondary structure and to monitor ligand interactions and conformational changes involving individual bases of RNA (Peattie and Gilbert 1980; Ehresmann et al. 1987; Stern et al. 1988; Hanson et al. 2005). For instance, tRNAs, initiation factors, elongation factors, and most antibiotics that bind to the ribosome have been shown to interact with ribosomal RNA, and their binding sites have been mapped using chemical probing (Moazed and Noller 1986, 1987; Moazed et al. 1988, 1995). More recently, ligand binding and the function of riboswitches have been characterized using in-line probing assays (Winkler et al. 2003; Barrick et al. 2004; Welz and Breaker 2007) or chemical probing (Hanson et al. 2005). In order to test our fluorescent primer extension method, we chose to observe binding of a well-characterized antibiotic to a 30S ribosomal subunit. Aminoglycosides cause decreases in translational accuracy and inhibit translocation (Davies et al. 1965) by binding to a conserved sequence of rRNA that is near the site of codon–anticodon recognition in the aminoacyl–tRNA site (A site) of 30S subunits (Moazed and Noller 1987; Fourmy et al 1996; Carter et al. 2000; Vicens and Westhof 2001). It has been shown previously that in the presence of 10 μM paromomycin, residues G1405, A1408, and G1494 were strongly protected from chemical modification by DMS, whereas G1491 and G1497 were weakly protected, with an estimated Kd of 0.2 μM (25°C) (Recht et al. 1996). Binding of paromomycin to 30S subunits was assayed by chemical probing with dimethyl sulfate (DMS) and the changes of reactivity of nucleotides were monitored by FPE. A strong footprint is observed at G1494(N7) (Fig. 3) and A1408(N1) in the presence of 10 μM paromomycin. This figure can be compared directly with previous studies that used radioactivity (Yoshizawa et al. 1998; Recht et al. 1999). Both methods, fluorescence and radioactivity, give similar results. Primer extension reactions, separation of cDNAs by PAGE, and fluorescent imaging of the gel were performed within 1 d. The results show that RNA–ligand interactions can be readily monitored using FPE with sensitivity comparable to methods using radioactivity.
FIGURE 3.
DMS probing reactions on 30S ribosomal subunits revealed with IRD-700. (Lane 1) A control reaction with no DMS added; (lane 2) a DMS probing reaction in the absence of paromomycin. Subunits were present at a concentration of 100 nM in all reactions. (Lanes 3–6) are reactions in the presence of 0.01, 1, 2.5, and 10 μM paromomycin, respectively. Nucleotides were identified by comparison with the dideoxy sequencing reactions (lanes U,G,C,A). Bands corresponding to nucleotide G1494 are indicated.
Poisoned primer extension assay
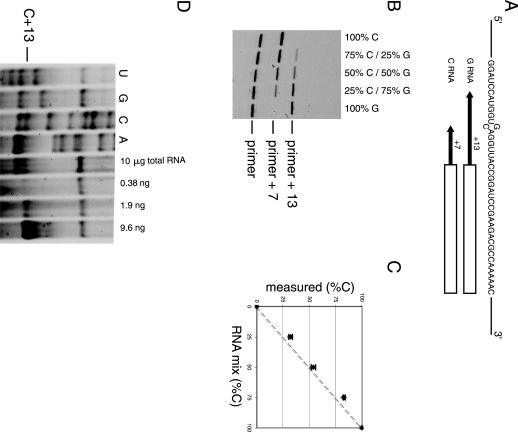
Poisoned primer extension (PPE) assay is used to measure the efficiency of RNA editing and to investigate mutant population in the RNA (Dabiri et al. 1996; Yoshizawa et al. 1999; Roberson and Rosenthal 2006). The assay is based on using a polymerase to extend a labeled primer through the mutation (or edited) site in the presence of three deoxynucleotide triphosphates and one dideoxynucleotide triphosphate. The dideoxynucleotide contains the base complementary to the mutation. If there is a mutation, the extension stops at the mutation site. On wild-type RNA, reverse transcriptase runs through this site and terminates further downstream at the next nucleotide with the same identity as at the mutated site. The population of the mixture can be determined by analyzing the resulting extension products on polyacrylamide gels. A PPE assay was performed using an IRD-700-labeled primer. We prepared a 2.5-kb RNA fragment containing a single G-to-C mutation at position 738. The primer was designed to hybridize at a 7 nt distance on the 3′ side of the mutation site. DideoxyGTP was used for the primer extension reaction. The C-RNA gives a product with 7 nt added to the primer, whereas 13 nt are added for G-RNA (Fig. 4A). Various mixtures of these two RNAs were prepared to verify if the assay is quantitative (Fig. 4B). Band intensities were quantified and plotted against the theoretical ratio of the RNAs (Fig. 4C). The assay was performed using 5 ng of RNA (6 fmol), which is comparable to the quantity used in previously published protocol with the 32P-labeled primer (Dabiri et al. 1996). The discrepancy between predicted and actual results was within 8%. The results were highly reproducible with a standard deviation of less than ±2% for each point for three independent experiments.
FIGURE 4.
Poisoned primer extension assay using IRD-700-labeled primer. (A) Sequence around the mutation site is shown. The box indicates the sequence complementary to the primer (23 mer) and the arrows indicate the extension until the first instance of C for each RNA. (B) Fluorescence scan of a polyacrylamide gel analyzing primer extension products of various G- and C-RNA mixtures. The primer, +7 (for C-RNA), and +13 (for G-RNA) products are indicated. (C) Population of C-RNA measured plotted against theoretical concentration. The dotted line represents the expected results if known and measured values are identical. The error bars represent the standard deviation of three independent experiments. (D) Polyacrylamide gel analyzing primer extension products of the mRNA. The four lanes on the left are sequencing ladders of in vitro transcribed mRNA. The position of the first C encountered by the reverse transcriptase in presence of ddGTP is indicated (C+13). The pause at C+13 was observed with 10 μg of total yeast RNA extract. The three lanes on the right show the pause at C+13 obtained on different quantities of in vitro transcribed RNA. These three lanes served as a crude calibration to estimate the intensity of the C+13 band obtained with 10 μg of total yeast RNA extract.
The PPE method can also be used to detect relatively low abundant mRNA in cell extracts. Total RNA from Saccharomyces cerevisiae cells was extracted, and the PPE method was applied to detect the presence of the mRNA from the open reading frame YMR116C (38 copies/cell) (Velculescu et al. 1997). The result of a PPE experiment using a DNA primer labeled at its 5′ end with IRD-700 dye and complementary to bases 940–960 of YMR116C mRNA is shown in Figure 4D. In the presence of three deoxynucleotide triphosphates (dATP, dCTP, and dUTP), and one dideoxynucleotide triphosphate (ddGTP), a pause of the reverse transcriptase is observed at the first C nucleotide (C+13) after the hybridization site for the primer (Fig. 4D). A pause was observed as expected at position A+7 with ddUTP confirming that the correct sequence of the mRNA is probed. Quantification of the fluorescent band leads to a value of ∼10 copies of mRNA per cell, which is in the same range as the previous value of 38 copies per cell (Velculescu et al. 1997).
Protein–RNA interactions monitored by fluorescent mobility shift assays (EMSA)
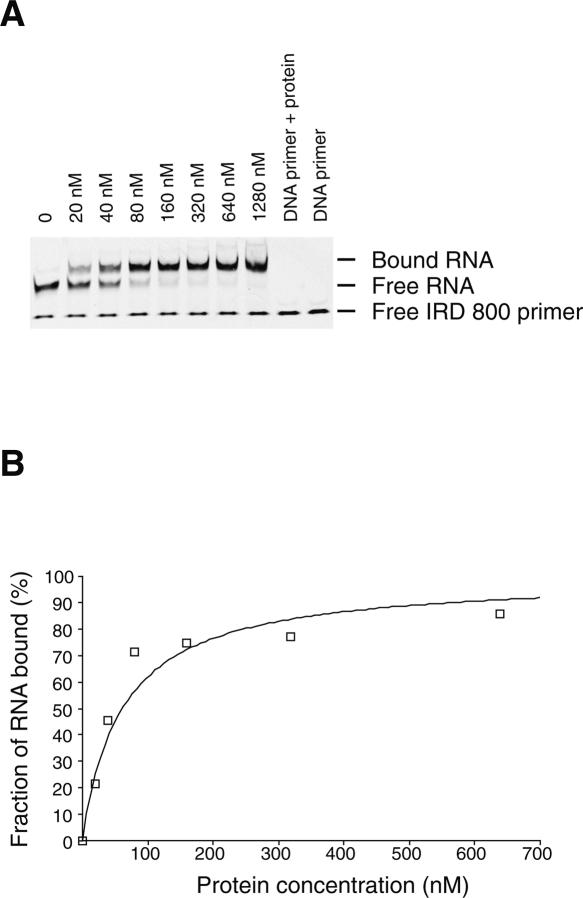
In order to test the use of fluorescently labeled primers for monitoring protein–RNA interactions by EMSA we have chosen the well-studied interaction between the mRNA binding domain of elongation factor SelB and its mRNA target: the selenocysteine insertion sequence (SECIS) (Fourmy et al. 2002; Yoshizawa et al. 2005). Winged-helix motifs specifically recognize the stem–loop structure formed by SECIS with high affinity (0.2 nM). Precise measurement of the apparent dissociation constant requires a concentration of RNA below the Kd (Batey and Williamson 1996). The RNA is detected by hybridization of an IRD-labeled primer to a 3′ single-stranded extension. The primer concentration was adjusted to a 10-fold excess with respect to the RNA concentration in order to maximize RNA detection. We found that the limit of detection of RNA by an IRD-labeled DNA primer by PAGE is 1 nM. However, the signal is more reliable at 10 nM of RNA and slightly better with IRD-700 rather than IRD-800. Therefore, we chose to measure the apparent dissociation constant for the SECIS RNA sequence for binding to a mutant (R510A) SelB C-terminal domain that has a Kd in the 100 nM range (Soler et al. 2007) using EMSA (Fig. 5). The fluorescent primer is hybridized to a 3′ 17-nt extension of the RNA prior to formation of the protein–RNA complex. The three-dimensional structure of the hairpin is critical for high affinity (Soler et al. 2007). Before the protein binding assays, the RNA was placed at 95°C for 5 min in the presence of the DNA primer and cooled on ice for 10 min in order to fold the RNA. Titration of the SelB domain by SECIS RNA yielded a complex of lower electrophoretic mobility than the unbound RNA (Fig. 5), as previously observed with the use of a 32P-labeled fdhF SECIS RNA (Fourmy et al. 2002). To determine the apparent dissociation constant for this RNA–protein interaction, the fluorescence intensity of the bands in the free and bound RNA was quantified, and the binding data were fit to an equation that describes a simple bimolecular equilibrium (Fig. 5B). The apparent Kd of 62 (±8) nM measured is similar to the one obtained by the previous radioactivity method (Soler et al. 2007). In order to evaluate the limits of this method, we measured the affinity of the wild-type SelB fragment with an RNA concentration of 1 nM. Free and bound fractions of RNA were measured with the IRD-700 version of the primer. The value of the apparent Kd is 1 nM, slightly higher than the previous value of 0.2 nM (Fourmy et al. 2002). Even though the limit of detection of the RNA at 1 nM prevents an exact measurement of the apparent Kd, this method can still provide useful information for protein–RNA interactions. Thus, by fluorescent EMSA, we can readily monitor protein–RNA interactions and precisely measure apparent dissociation constants as low as 10 nM. The fact that the apparent Kd measured for the SECIS RNA containing a 3′ extension hybridized to the fluorescent DNA oligonucleotide is very similar to the Kd for the original SECIS hairpin demonstrates that the 3′ extension does not affect the overall structure of the hairpin. This result is consistent with the crystal structure of the SelB C-terminal domain in complex with the SECIS RNA stem–loop, where the protein only contacts the upper region of the stem–loop and not the 3′ end (Soler et al. 2007). Thus, here the 3′ extension does not alter the protein–RNA interaction and provides an easy and inexpensive method to monitor the interaction. However, in some RNA molecules, adding a 3′ extension may alter its conformation and affect its affinity for the protein. An alternative approach, but more costly, is to synthesize, in-house, a 5′-labeled RNA using IRDye phosphoramidites. This approach has the potential of improving the detection limit of the RNA (down to 0.2 fmol), since there is no risk of dissociation of the dye from the RNA during gel electrophoresis. This method would lead to a precise determination of apparent Kd in the nanomolar range or slightly below.
FIGURE 5.
(A) Titration of SelB (R510A) domain into IRD-800-labeled fdhF SECIS RNA. This titration was performed at 4°C and yields an apparent dissociation constant of ∼62 (±8) nM. (B) Graph showing the fraction of bound fdhF RNA as a function of increasing concentration of SelB (R510A) fragment.
SHAPE experiment on 30S ribosomal subunit
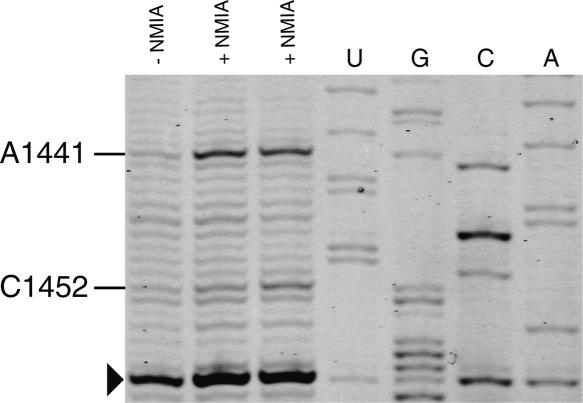
Recently, a quantitative approach has been developed involving selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE) to map and distinguish fine differences in structures for RNA molecules at single nucleotide resolution (Merino et al. 2005; Mortimer and Weeks 2007). This method has been successfully applied to reveal the structural changes that occur during the crystallization process of the P4-P6ΔC209 domain Tetrahymena Thermophila group I intron (Vicens et al. 2007). Here we show that this approach can be used to rapidly probe the rRNA structure in ribosomal subunits using a primer extension with IRD-700-labeled primers. Selective acylation of the ribose 2′-hydroxyl position by N-methylisatoic anhydride (NMIA) is exquisitely sensitive to local nucleotide flexibility. The reactivity of nucleotides from a segment of helix 44 in 16S rRNA is shown in Figure 6. This helix is found at the ribosomal subunit interface in the 70S ribosome (Schuwirth et al. 2005; Selmer et al. 2006). The SHAPE experiment accurately reports the known structure of rRNA. Nucleotide C1452 has medium reactivity with NMIA that correlates with a C2′ endo conformation in the context of a UUCG tetraloop (Cheong et al. 1990). Nucleotide A1441 is found highly reactive and is bulged out from helix 44 in the E. coli 30S subunit structure. It interacts with the minor groove edge of the G1461–U1440 pair in helix 44 (Schuwirth et al. 2005). The SHAPE experiment combined with fluorescent IRD-700 primers for reverse transcription allows fast chemical probing of the RNA structure at single nucleotide resolution.
FIGURE 6.
Example of a SHAPE experiment performed on 30S ribosomal subunits as visualized by PAGE with IRD-700. Reactive nucleotides in this region of H44 of 16S rRNA are indicated. Nucleotides were identified by comparison with the dideoxy sequencing reactions (lanes U,G,C,A). The arrowhead indicates a nucleotide corresponding to a pause of the reverse transcriptase.
Conclusion
Radioactivity has been an essential tool for biochemical and structural investigation of RNA. For instance, chemical probing methods are largely used for mapping RNA secondary structure and to monitor ligand interactions and conformational changes involving individual bases of RNA (Peattie and Gilbert 1980; Ehresmann et al. 1987; Stern et al. 1988; Soukup and Breaker 1999; Hanson et al. 2005; Merino et al. 2005). However, until now, these methods required 33P or 32P labeling of the RNA or DNA primers. The recent development of fluorescent dyes and fluorescent imaging systems allows the substitution of radioactivity by fluorescent methodologies with similar sensitivity. Here, using infrared dyes and an infrared fluorescence imaging system, we demonstrated fast and sensitive investigation of RNA sequencing, chemical probing, protein–RNA, and ligand interactions. The speed and ease of these approaches are advantages over prior methods that use hazardous radioisotopes. Structural and biochemical investigations of RNA should benefit from the use of these fluorescent methodologies.
MATERIALS AND METHODS
Preparation of yeast ribosomal RNAs
S. cerevisiae cells grown at the exponential phase were recovered by centrifugation at 5000 rpm for 5 min (rotor JA 91,000, Beckman). Cell pellets were resuspended in the lysis buffer (20 mM Hepes-KOH at pH 7.6, 100 mM KCl, 10 mM MgCl2, and 1 mM DTT) and broken by passing through a cell disruptor equipment (BazicZ, CellD) at 1400 bar and 4°C. Cell debris were removed by centrifugation at 15,000 rpm for 30 min (rotor 70Ti, Beckman), and for another 3 h in the presence of 20% glycerol. Sucrose (final concentration at 0.5 M) was added to the cell extract (supernatant), following ultracentrifugation at 31,000 rpm for 4 h (rotor 70 Ti, Beckman). The resulting pellet (ribosomes) was resuspended in a buffer containing 20 mM Hepes-KOH (pH 7.6), 100 mM KCl, 10 mM MgCl2, and 7 mM β-Mercaptoethanol. Three phenol extractions (in the presence of 1% of SDS) were followed by two chloroform extractions in order to isolate rRNAs. The purity and the size of rRNAs were verified by agarose gel electrophoresis.
Extraction of total RNAs
S. cerevisiae (S288C) cells grown at the exponential phase were recovered and washed twice. Cell pellets (∼200 mg) were resuspended in the extraction buffer (∼1.2 mL), containing 10 mM Tris-HCl (pH 7.5), 10 mM EDTA (pH 8.0), and 0.5% SDS. One volume of phenol solution was added and the mixture was incubated at 65°C for 1 h under strong agitation (vortex ∼1400 rpm). After leaving the reaction tube on ice for 5 min, two phenol extractions and one chloroform extraction were carried out. The resulting total RNAs were precipitated by ethanol and dissolved in 200 μL of RNase-free water. The RNA (150 μL) was then further purified using two spin columns from RNeasyMiniKit (QIAGEN). The final yield of RNA was 100 μg.
RNA sequencing using radioactive primer extension
Primer extension was performed using adenosine 5′-triphosphate [α-32P] (PerkinElmer) and reverse transcriptase SuperScript II (Invitrogen). The experimental procedure was carried out as described previously (Yoshizawa et al. 1998). A total of 0.6 pmol of 16S rRNA (E. coli) and 10 pmol of the primer complementary to nucleotides 1530–1509 of 16S rRNA was employed for each reaction. The final samples were dissolved with 10 μL of loading buffer, containing 7 M urea, bromophenol blue, and xylene cyanol blue. Then, 2.5 μL of the samples were loaded on 8% polyacrylamide gels containing 7 M urea (thickness 0.4 mm, width 300 mm, length 340 mm) for separation. After electrophoresis, the gel was dried with a piece of 3MM Whatman filter paper, and subsequently exposed to a PhosphorImager screen overnight. The radioactivity was detected using a PhosphorImager Storm (Molecular Dynamics).
Chemical probing of E. coli 30S ribosomal subunits
Modification reactions (100 μL) with 30S subunits (10 pmol) were performed in a buffer containing 80 mM potassium cacodylate (pH 7.2), 100 mM ammonium chloride, 20 mM magnesium chloride, 1 mM dithiothreitol, and 0.5 mM EDTA, as previously described (Yoshizawa et al. 1998). Paromomycin was added and modification was performed by addition of DMS (15 μL of a 1/10 dilution in ethanol) followed by incubation at room temperature for 10 min and on ice for 10 min. Reactions were stopped by ethanol precipitation. Sodium borohydride reduction and aniline-induced strand scission was performed as described (Recht et al. 1996). Modified RNA was resuspended in 10 μL of 1 M Tris-HCl (pH 8.2). Upon addition of 10 μL of freshly prepared 0.2 M NaBH4, the samples were incubated on ice in the dark for 30 min. The reaction was quenched by addition of 100 μL of 0.4 M sodium acetate, followed by ethanol precipitation. Pellets were dissolved in 20 μL of 1.0 M aniline/acetate (pH 4.5), followed by incubation in the dark for 20 min at 60°C. The reaction was quenched by addition of 100 μL of 0.4 M sodium acetate and 100 μL of phenol:chloroform:isoamyl alcohol (25:24:1), followed by vigorous mixing and centrifugation. The RNA was concentrated by ethanol precipitation of the aqueous phase and pellets were washed with 100 μL of cold 70% ethanol.
Fluorescent primer extension
IRD-700 and IRD-800 fluorescent DNA primers were purchased from MWG biotech. Primers were dissolved in 50 mM Tris-HCl (pH 8.1) and kept frozen in small aliquots at a concentration of 100 μM. Primers were all 5′-end labeled. FPE for sequencing of yeast rRNAs was performed using fluorescent primers, either IRD-700 or IRD-800 labeled, complementary to nucleotides 134–158 of 5.8S rRNA, and nucleotides 168–187, 343–362, and 478–498 of 25S rRNA. Primer extension of E. coli 16S rRNA (Stern et al. 1988) was performed using an IRD-700 fluorescent DNA primer that is complementary to nucleotides 1530–1509 of 16S rRNA. Total yeast rRNAs (1 μg) or modified 16S E. coli rRNA (2.5 μL, 0.6 pmol) were added to the fluorescent reverse transcription primer (10 pmol) in hybridization buffer (25 mM Hepes-KOH at pH 7.0, 50 mM KCl), heated to 90°C (1–2 min), and slowly cooled to 45°C. Reverse transcription (RT) reactions were performed in 50 mM Tris-HCl (pH 8.3), 75 mM KCl, 3 mM MgCl2, and 10 mM DTT with 0.25 mM each dNTP. SuperScript II (Invitrogen, 1 μL 200 units) was added and reactions were incubated at 45°C for 40 min. For sequencing samples, 1.25 mM of ddATP, ddCTP, ddGTP, or ddTTP were supplied individually to the RT reactions of each sample. Reactions were stopped by ethanol precipitation. cDNA samples were dissolved in 10 μL of loading dye solution containing 7 M urea, 1X TBE, and bromophenol blue (xylene cyanol blue is omitted as it is detected at 700 nm). A small aliquot (1.5–3 μL) of each sample was loaded on an 8% polyacrylamide gel for separation of cDNAs. The gel is transferred from the electrophoresis glass plates using a rigid plastic sheet and covered with plastic wrap. The gel is then applied to the infrared imaging system Odyssey (LI-COR Biosciences) with the plastic wrap contacting the imaging system. The plastic sheet is then removed. Bands were detected and quantified directly on the infrared imaging system.
Poisoned primer extension
RNAs (2.5 kb) containing a single base difference (G or C) at position 738 were transcribed from PCR amplified templates using T7 RNA polymerase. The transcripts were phenol extracted, ethanol precipitated, and load onto a Sephadex G50 (sigma) column (5.5 mL) to remove the remaining NTPs. RNA (5 ng) was hybridized with 11.2 fmols of 5′ IRD700 modified DNA primer (23 mer) in 2.3 μL of buffer containing 50 mM Hepes-KOH (pH7.0) and 100 mM KCl by heating at 90°C for 1 min followed by slow cooling to 45°C. Primer extension was performed in 50 mM Tris-HCl (pH 8.3), 75 mM KCl, 3 mM MgCl2, and 10 mM DTT with 0.5 mM each of dATP, dCTP, dTTP, and dideoxyGTP. SuperScript II (100 U) was added and reactions were incubated at 45°C for 25 min. The reaction mixtures were ethanol precipitated and analyzed on a 20% 7 M urea polyacrylamide gel.
Gel mobility shift analysis.
The complex between the SelB fragment and SECIS RNA was observed as a mobility shift in a nondenaturing polyacrylamide gel as previously described (Fourmy et al. 2002). Prior to complex formation, a fluorescent DNA primer with a sequence complementary to a 3′ 17-nt extension (5′-GGUUGGCGUGGCUCGCG-3′) on the RNA was hybridized to SECIS RNA (1/1 ratio) by heating to 95°C for 5 min and cooled on ice for 10 min in 100 mM Tris-HCl (pH7.0). A constant concentration of fluorescent DNA–RNA hybrid (10 nM) was incubated with various concentrations of protein in 15 μL of 10 mM sodium phosphate (pH6.0), 100 mM NaCl, 0.1 mM EDTA, 1 mM DTT, 2 μM competitor RNA (24 nt), and 0.01% Nonidet P-40. The reaction mixture was incubated at room temperature for 10 min and then on ice for 10 min; 30% glycerol dye (bromophenol blue) was added and the complex was separated from the free RNA by electrophoresis in 10% polyacrylamide (acrylamide:bisacrylamide 29:1)-1X TBE gels and quantitated using an infrared imaging system Odyssey (LI-COR Biosciences). Data were fit using the MC-Fit program (Dardel 1994) to obtain apparent dissociation constants.
SHAPE experiment
30S ribosomal subunits (10 pmol; 45 μL) were treated with NMIA (Invitrogen, 5 μL, 130 mM in anhydrous DMSO, 35°C) and allowed to react for 54 min as previously described (Merino et al. 2005). Control reactions contained 5 μL of DMSO in place of NMIA. Modified 16S rRNA was recovered from 30S subunits by phenol extraction as described above. Modified RNA was subjected to FPE as described above.
ACKNOWLEDGMENTS
We thank Cyril Gaudin for providing the 30S ribosomal subunit and Nicolas Soler for providing the R510A SelB domain and SECIS RNA. We are also grateful to Jean-Claude Drapier for helpful discussions and to Koen Visscher for critical reading of the manuscript. This work was supported in part by a grant from ANR young investigator (JC05-50413) to S.Y. and D.F. B.-W.Y. was supported by a fellowship from the Institut de Chimie des Substances Naturelles.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.637907.
REFERENCES
- Barrick, J.E., Corbino, K.A., Winkler, W.C., Nahvi, A., Mandal, M., Collins, J., Lee, M., Roth, A., Sudarsan, N., Jona, I., et al. New RNA motifs suggest an expanded scope for riboswitches in bacterial genetic control. Proc. Natl. Acad. Sci. 2004;101:6421–6426. doi: 10.1073/pnas.0308014101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batey, R.T., Williamson, J.R. Interaction of the Bacillus stearothermophilus ribosomal protein S15 with 16 S rRNA: II. Specificity determinants of RNA–protein recognition. J. Mol. Biol. 1996;261:550–567. doi: 10.1006/jmbi.1996.0482. [DOI] [PubMed] [Google Scholar]
- Carter, A.P., Clemons, W.M., Brodersen, D.E., Morgan-Warren, R.J., Wimberly, B.T., Ramakrishnan, V. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature. 2000;407:340–348. doi: 10.1038/35030019. [DOI] [PubMed] [Google Scholar]
- Cheong, C., Varani, G., Tinoco, I.J. Solution structure of an unusually stable RNA hairpin, 5′GGAC(UUCG)GUCC. Nature. 1990;346:680–682. doi: 10.1038/346680a0. [DOI] [PubMed] [Google Scholar]
- Dabiri, G.A., Lai, F., Drakas, R.A., Nishikura, K. Editing of the GLuR-B ion channel RNA in vitro by recombinant double-stranded RNA adenosine deaminase. EMBO J. 1996;15:34–45. [PMC free article] [PubMed] [Google Scholar]
- Dardel, F. MC-Fit: Using Monte Carlo methods to get accurate confidence limits on enzyme parameters. Comput. Appl. Biosci. 1994;10:273–275. doi: 10.1093/bioinformatics/10.3.273. [DOI] [PubMed] [Google Scholar]
- Davies, J., Gorini, L., Davis, B.D. Misreading of RNA codewords induced by aminoglycoside antibiotics. Mol. Pharmacol. 1965;1:93–106. [PubMed] [Google Scholar]
- Ehresmann, C., Baudin, F., Mougel, M., Romby, P., Ebel, J.P., Ehresmann, B. Probing the structure of RNAs in solution. Nucleic Acids Res. 1987;15:9109–9128. doi: 10.1093/nar/15.22.9109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourmy, D., Recht, M.I., Blanchard, S.C., Puglisi, J.D. Structure of the A site of E. coli 16S rRNA complexed with an aminoglycoside antibiotic. Science. 1996;274:1367–1371. doi: 10.1126/science.274.5291.1367. [DOI] [PubMed] [Google Scholar]
- Fourmy, D., Guittet, E., Yoshizawa, S. Structure of prokaryotic SECIS mRNA hairpin and its interaction with elongation factor SelB. J. Mol. Biol. 2002;324:137–150. doi: 10.1016/s0022-2836(02)01030-6. [DOI] [PubMed] [Google Scholar]
- Hanson, S., Bauer, G., Fink, B., Suess, B. Molecular analysis of a synthetic tetracycline-binding riboswitch. RNA. 2005;11:503–511. doi: 10.1261/rna.7251305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machida, M., Kamio, H., Sorensen, D. Long-range and highly sensitive DNase I footprinting by an automated infrared DNA sequencer. Biotechniques. 1997;23:300–303. doi: 10.2144/97232st08. [DOI] [PubMed] [Google Scholar]
- Merino, E.J., Wilkinson, K.A., Coughlan, J.L., Weeks, K.M. RNA structure analysis at single nucleotide resolution by selective 2′-hydroxyl acylation and primer extension (SHAPE) J. Am. Chem. Soc. 2005;127:4223–4231. doi: 10.1021/ja043822v. [DOI] [PubMed] [Google Scholar]
- Middendorf, L.R., Bruce, J.C., Bruce, R.C., Eckles, R.D., Grone, D.L., Roemer, S.C., Sloniker, G.D., Steffens, D.L., Sutter, S.L., Brumbaugh, J.A., et al. Continuous, on-line DNA sequencing using a versatile infrared laser scanner/electrophoresis apparatus. Electrophoresis. 1992;13:487–494. doi: 10.1002/elps.11501301103. [DOI] [PubMed] [Google Scholar]
- Moazed, D., Noller, H.F. Transfer RNA shields specific nucleotides in 16S ribosomal RNA from attack by chemical probes. Cell. 1986;47:985–994. doi: 10.1016/0092-8674(86)90813-5. [DOI] [PubMed] [Google Scholar]
- Moazed, D., Noller, H.F. Interaction of antibiotics with functional sites in 16S ribosomal RNA. Nature. 1987;327:389–394. doi: 10.1038/327389a0. [DOI] [PubMed] [Google Scholar]
- Moazed, D., Robertson, J.M., Noller, H.F. Interaction of elongation factors EF-G and EF-Tu with a conserved loop in 23S RNA. Nature. 1988;334:362–364. doi: 10.1038/334362a0. [DOI] [PubMed] [Google Scholar]
- Moazed, D., Samaha, R.R., Gualerzi, C., Noller, H.F. Specific protection of 16S rRNA by translational initiation factors. J. Mol. Biol. 1995;248:207–210. doi: 10.1016/s0022-2836(95)80042-5. [DOI] [PubMed] [Google Scholar]
- Mortimer, S.A., Weeks, K.M. A fast-acting reagent for accurate analysis of RNA secondary and tertiary structure by SHAPE chemistry. J. Am. Chem. Soc. 2007;129:4144–4145. doi: 10.1021/ja0704028. [DOI] [PubMed] [Google Scholar]
- Peattie, D.A., Gilbert, W. Chemical probes for higher-order structure in RNA. Proc. Natl. Acad. Sci. 1980;77:4679–4682. doi: 10.1073/pnas.77.8.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recht, M.I., Fourmy, D., Blanchard, S.C., Dahlquist, K.D., Puglisi, J.D. RNA sequence determinants for aminoglycoside binding to an A-site rRNA model oligonucleotide. J. Mol. Biol. 1996;262:421–436. doi: 10.1006/jmbi.1996.0526. [DOI] [PubMed] [Google Scholar]
- Recht, M.I., Douthwaite, S., Puglisi, J.D. Basis for prokaryotic specificity of action of aminoglycoside antibiotics. EMBO J. 1999;18:3133–3138. doi: 10.1093/emboj/18.11.3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberson, L.M., Rosenthal, J.J. An accurate fluorescent assay for quantifying the extent of RNA editing. RNA. 2006;12:1907–1912. doi: 10.1261/rna.166906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuwirth, B.S., Borovinskaya, M.A., Hau, C.W., Zhang, W., Vila-Sanjurjo, A., Holton, J.M., Cate, J.H. Structures of the bacterial ribosome at 3.5 Å resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- Selmer, M., Dunham, C.M., Murphy, F.V.T., Weixlbaumer, A., Petry, S., Kelley, A.C., Weir, J.R., Ramakrishnan, V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- Soler, N., Fourmy, D., Yoshizawa, S. Structural insight into a molecular switch in tandem winged-helix motifs from elongation factor SelB. J. Mol. Biol. 2007;370:728–741. doi: 10.1016/j.jmb.2007.05.001. [DOI] [PubMed] [Google Scholar]
- Soukup, G.A., Breaker, R.R. Relationship between internucleotide linkage geometry and the stability of RNA. RNA. 1999;5:1308–1325. doi: 10.1017/s1355838299990891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern, S., Moazed, D., Noller, H.F. Structural analysis of RNA using chemical and enzymatic probing monitored by primer extension. Methods Enzymol. 1988;164:481–489. doi: 10.1016/s0076-6879(88)64064-x. [DOI] [PubMed] [Google Scholar]
- Velculescu, V.E., Zhang, L., Zhou, W., Vogelstein, J., Basrai, M.A., Bassett, D.E., Hieter, P., Vogelstein, B., Kinzler, K.W. Characterization of the yeast transcriptome. Cell. 1997;88:243–251. doi: 10.1016/s0092-8674(00)81845-0. [DOI] [PubMed] [Google Scholar]
- Vicens, Q., Westhof, E. Crystal structure of paromomycin docked into the eubacterial ribosomal decoding A site. Structure. 2001;9:647–658. doi: 10.1016/s0969-2126(01)00629-3. [DOI] [PubMed] [Google Scholar]
- Vicens, Q., Gooding, A.R., Laederach, A., Cech, T.R. Local RNA structural changes induced by crystallization are revealed by SHAPE. RNA. 2007;13:536–548. doi: 10.1261/rna.400207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welz, R., Breaker, R.R. Ligand binding and gene control characteristics of tandem riboswitches in Bacillus anthracis . RNA. 2007;13:573–582. doi: 10.1261/rna.407707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, D.C., Soper, S.A. Ultrasensitive near-IR fluorescence detection for capillary gel electrophoresis and DNA sequencing applications. Anal. Chem. 1995;67:3427–3432. doi: 10.1021/ac00115a010. [DOI] [PubMed] [Google Scholar]
- Winkler, W.C., Nahvi, A., Sudarsan, N., Barrick, J.E., Breaker, R.R. An mRNA structure that controls gene expression by binding S-adenosylmethionine. Nat. Struct. Biol. 2003;10:701–707. doi: 10.1038/nsb967. [DOI] [PubMed] [Google Scholar]
- Yoshizawa, S., Fourmy, D., Puglisi, J.D. Structural origins of gentamicin antibiotic action. EMBO J. 1998;17:6437–6448. doi: 10.1093/emboj/17.22.6437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizawa, S., Fourmy, D., Puglisi, J.D. Recognition of the codon–anticodon helix by ribosomal RNA. Science. 1999;285:1722–1725. doi: 10.1126/science.285.5434.1722. [DOI] [PubMed] [Google Scholar]
- Yoshizawa, S., Rasubala, L., Ose, T., Kohda, D., Fourmy, D., Maenaka, K. Structural basis for mRNA recognition by elongation factor SelB. Nat. Struct. Mol. Biol. 2005;12:198–203. doi: 10.1038/nsmb890. [DOI] [PubMed] [Google Scholar]