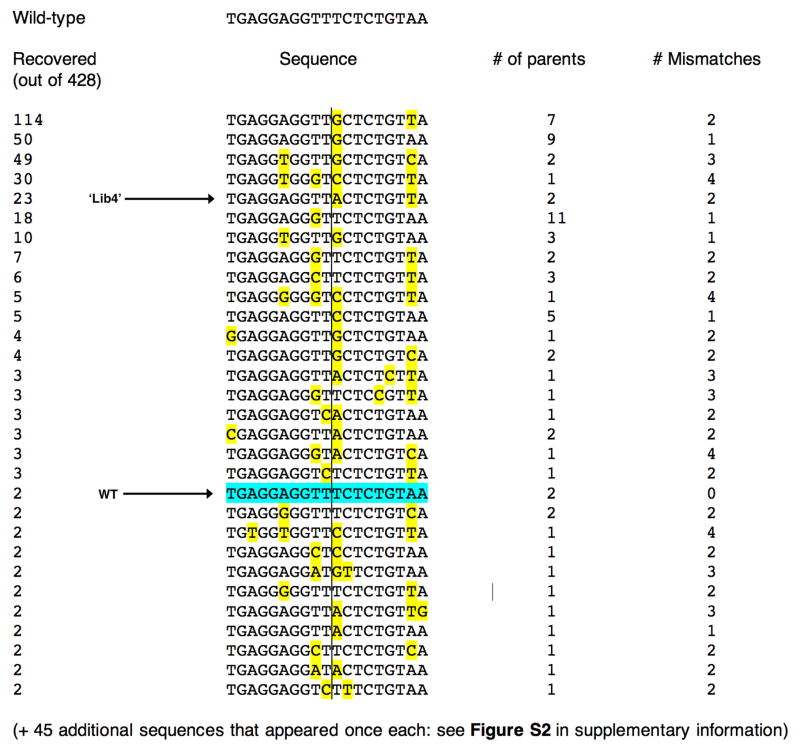
Figure 2. Target sites recovered in a specificity measurement screen of the I-AniI endonuclease.
In this experiments, 428 substrate targets were sequenced. The wild-type site was isolated twice in this screen and is indicated; individual mismatches in other sites, relative to the wild-type target, are highlighted in yellow. The center of the target site (dividing the left and right half-sites) is indicated with the vertical line. In addition to individual sequences of cleavable substrates, the number of individual parental clones for each target can be identified, based on unique sequences of randomized, addressible flanking bases. Out of 76 unique sequences, 31 appeared more than once, out of which half were generated from multiple parental site clones. The frequency at which individual sites were recovered did not correlate strongly with their performance in kinetic cleavage assays; however at least one site (‘Lib4’) is a much better substrate than the physiological, wild-type target sequence (see Figure 4).