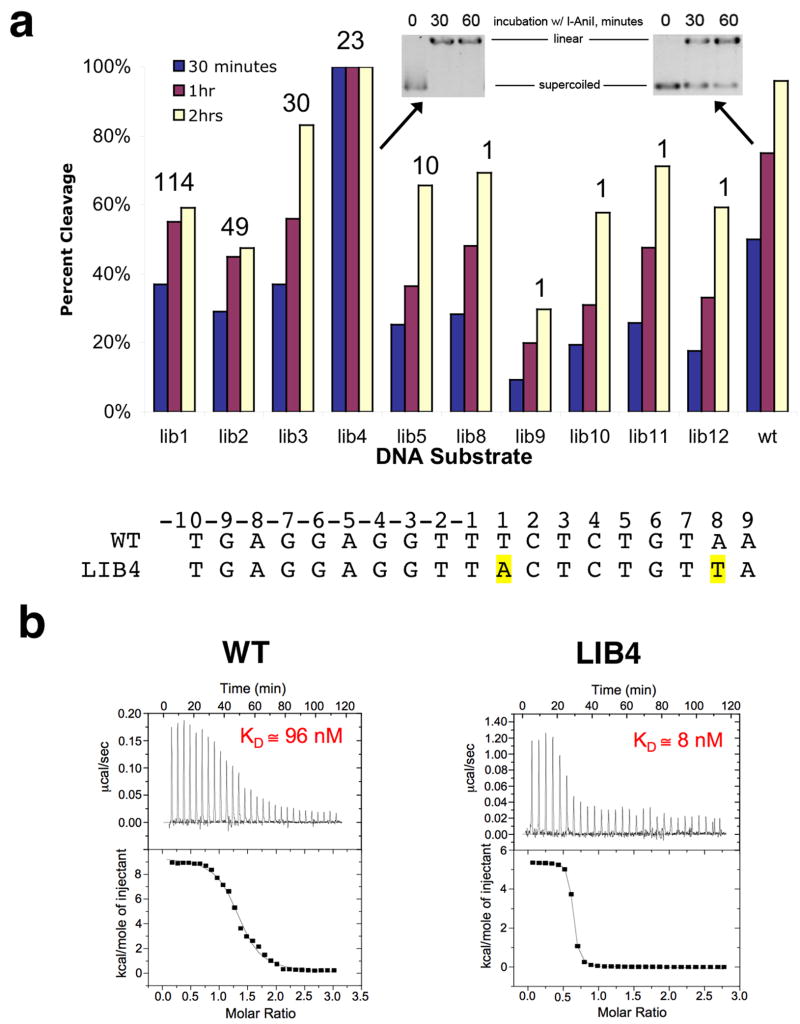
Figure 5. Relative behavior of isolated target sites as cleavage substrates for I-AniI.
Panel a: Kinetics of cleavage under reaction conditions as described in ‘methods’. Extent of cleavage quantitated at 30, 60, and 120 minutes. The numbers above the individual substrate bars correspond to the number of times each sequence was isolated in the screen, as shown in the first column of Figure 1. Note the complete rapid digestion of ‘Lib4’ relative to the wild-type target sequence. Panel b: Relative binding of wild-type and ‘Lib4’ substrates by the I-AniI endonuclease, determined by isothermal titration calorimetry. The Kd values are averages of multiple independent experiments conducted on separate days, which reproducibly indicate that the ‘Lib4’ sequence is bound approximately 10-fold more tightly than the wild-type sequence. While the estimated molar ratios of binding for the experiments shown in these panels are slightly above and below 1:1, respectively, the average of multiple runs in each case indicates a 1:1 binding stoichiometry of protein to DNA duplex. The variation in this value illustrated in this figure is typical of ITC runs with homing endonuclease/DNA systems.