Abstract
Growing evidence suggests that the β-amyloid (Aβ) peptides of Alzheimer's disease are generated in early endosomes and that small oligomers are the principal toxic species. We sought to understand whether and how the solution pH, which is more acidic in endosomes than the extracellular environment, affects the conformational processes of Aβ. Using constant pH molecular dynamics simulations of two model peptides, Aβ(1–28) and Aβ(10–42), we found that the folding landscape of Aβ is strongly modulated by pH and is most favorable for hydrophobically driven aggregation at pH 6. Thus, our theoretical findings substantiate the possibility that Aβ oligomers develop intracellularly before secretion into the extracellular milieu, where they may disrupt synaptic activity or act as seeds for plaque formation.
Keywords: β-turn, helix, pH-dependent, molecular dynamics, electrostatics
Protein aggregation and fibril formation have been implicated in a number of human diseases, including amyloidoses and neurodegenerative disorders such as Alzheimer's disease (AD) and Parkinson's disease (PD) that are most prevalent in aged populations. Although it is now widely known that the initial aggregation step for globular proteins involves the formation of a partially unfolded monomeric intermediate (1), little is understood for natively unfolded proteins such as the β-amyloid (Aβ) peptides of AD and the α-synuclein protein of PD. To elucidate the conformational behavior of Aβ, fragments as well as the full-length (40 or 42 residues) monomers have been intensively studied by spectroscopic techniques such as circular dichroism (CD) and solution NMR in both water and aqueous solutions of TFE, a cosolvent known to stabilize local hydrogen bonding in peptides with intrinsic helix propensity (2). In water, Aβ peptides have been characterized as mainly unfolded (3, 4) whereas in TFE solution, they display a pH-dependent helix profile with a minimum helix content centered around pH 5.5 (3). An early study showed that Aβ(1–28) forms mature fibrils only in the pH range between 3 and 8 (5). A recent kinetics experiment revealed that the full-length Aβ aggregates most rapidly at pH 4–5.7 (6). However, conformational transitions in the early aggregation step as well as its relationship to pH remain unknown. Understanding the role of environmental conditions such as solution pH in the oligomerization of Aβ is important, because increasing experimental data suggest that Aβ is cleaved from the amyloid precursor protein in early endosomes, which have a pH of ≈6 (7, 8) and that dimers and trimers are necessary and sufficient to disrupt synaptic activity (9, 10).
We have recently developed the continuous constant pH molecular dynamics (CPHMD) method (11, 12), which allows conformational dynamics to be microscopically coupled with protonation equilibria in the atomic-level simulation of biological macromolecules. Combined with a state-of-the-art conformational sampling protocol, the replica-exchange (REX) algorithm (13), and an improved continuum solvent model (14, 15), this method has opened a door to first-principles pKa prediction for proteins (16) and theoretical studies of pH-dependent conformational processes in biology (17, 18). We now use REX-CPHMD simulations to explore the folding of two model peptides, Aβ(1–28) and Aβ(10–42). Aβ(1–28) is a necessary model for investigating the pH-dependent conformational behavior of Aβ, because it encompasses all of the ionizable residues of the full-length peptide as well as the central hydrophobic cluster (CHC, Leu-17ValPhePheAla-21), which forms the core of the minimum fibril-forming sequence 16–22 (19). Our emphasis is on the physiologically relevant model, Aβ(10–42), which incorporates both the CHC and the C-terminal hydrophobic region (sequence 29–42). These two regions form β sheets in the proposed fibril models of Aβ(1–40) and Aβ(1–42) based on the solid-state and hydrogen–deuterium exchange NMR data (20, 21). To ensure conformational sampling convergence, we removed the first nine residues in the full-length Aβ, because they are disordered in fibrils (20, 21) and are likely not important for aggregation (20). Our simulation data reveal a conformational energy landscape for both peptides that is nonrandom and displays preferential states that reconcile and rationalize the existing structural and mutation data. Furthermore, modulation of the folding landscape by solution pH unveils a molecular mechanism for the early aggregation of Aβ.
Results and Discussion
pH-Dependent Helix Propensity.
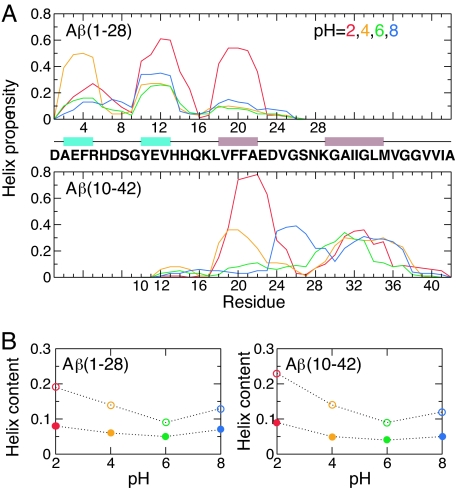
To provide a theoretical explanation for the disparity between the pH-dependent helix formation in TFE solution (3) and the mainly unfolded structure in water (3, 4), we performed a set of REX-CPHMD simulations for Aβ(1–28) at pH 2, 4, 6, and 8. Each 60-ns REX-CPHMD simulation was initiated from 16 replicas in a canonical helix structure (see Materials and Methods). The helix content of Aβ(1–28) rapidly decreases in the first 15 ns of simulation [supporting information (SI) Fig. 7]. Analysis of the equilibrium data reveals that the peptide is mainly unfolded with three regions, around residues 2–5, 10–13, and 18–22 displaying pronounced helix propensity (Fig. 1A). We denote these regions as “nascent” helices following the terminology introduced by Dyson et al. (2) for describing the induction of helices in natively unfolded peptides with turn-like motifs in the presence of organic cosolvent. The helix propensity of these segments depends on pH and reaches its lowest value at pH 6 and its highest value at pH 2. We note that the CHC region is particularly strongly modulated by pH (Fig. 1A). To facilitate a direct comparison with CD data, we obtained helix content based on the theoretical mean residue ellipticity at 222 nm. In agreement with the CD measurement (3), computed helix content is below 10% at all pH and shows a weak pH dependence (Fig. 1B). The contrast between the negligible total helix content and the appreciable propensity at the residue level can be readily understood because CD measurements are insensitive to short nascent helical fragments (see Materials and Methods).
Fig. 1.
pH-dependent helix formation. (A) Residue-based helix propensity of Aβ(1–28) and Aβ(10–42) computed from simulations at pH 2 (red), 4 (orange), 6 (green), and 8 (blue). Nascent helices containing ionizable residues are depicted in cyan, and those in the CHC and C-terminal hydrophobic regions are depicted in brown. (B) Total helix content as a function of pH computed as the percentage of helical residues (open symbols) and from the theoretical mean residue ellipticity at 222 nm (filled symbols).
To verify that pH-dependent nascent helix formation is also present in the full-length Aβ, and to ensure that this is not a biased result because of the helical starting structure, we carried out REX-CPHMD simulations of Aβ(10–42) starting from a fully extended structure. The equilibrium data shows that the peptide has two nascent helices, around 18–22 and 29–35 (Fig. 1A). Both regions are involved in β-sheet formation in the fibril (20, 21). Although the propensity for the first helix diminishes at pH 6 and 8, consistent with that in Aβ(1–28), the 29–35 region remains helical under all pH conditions, which can be attributed to the lack of adjacent residues that titrate in this pH range, 2–8. The calculated total helix content of Aβ(10–42) as well as its pH profile is almost identical to that of Aβ(1–28) (Fig. 1B). Aβ peptides were observed to undergo an α-helix-to-β-sheet transition with a bell-shaped pH profile in TFE solution (3). Because the β-sheet formation involves the CHC and C-terminal hydrophobic residues in the full-length Aβ, our data suggest that the CHC region is, in fact, responsible for the pH-dependent α-to-β transition. Thus, our data support the notion of a discordant region that has the highest predisposition for the α-to-β interconversion (22). Mutations that increase the helicity of this region would prevent the α-to-β conversion and consequently abolish fibril formation (23).
pH-Dependent Solvent Exposure.
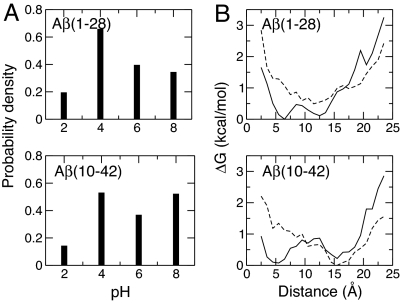
A solution NMR study of Aβ(10–35) revealed that the CHC residues form a hydrophobic patch on the protein surface (24). A most recent study using Bis-ANS fluorescence suggested that residues 13–21 and 30–36 are solvent-exposed (25). Because the nonspecific association of solvent-exposed hydrophobic residues represents a major driving force for protein aggregation (1), we hypothesized that the pH-dependent fibril formation (5), early aggregation kinetics (6), and deposition to AD authentic plaques (26) may be a result of the pH-modulated solvent exposure of the hydrophobic regions in Aβ. To test this hypothesis, we computed the solvent-accessible surface area using the equilibrium conformations of Aβ(1–28) and Aβ(10–42) at all pH. Fig. 2 shows that the CHC region in both Aβ(1–28) and Aβ(10–42) becomes more exposed as pH increases from 2 to 6 but more buried as pH is further increased to 8. In contrast, solvent exposure of the C-terminal hydrophobic region in Aβ(10–42) remains unchanged with respect to pH (data not shown). Thus, our data suggests that a pH-dependent exposure of the CHC region may help explain the observations that Aβ peptides form fibrils at pH 3–8 (5), aggregate most rapidly at pH 4–5.7 (6), and become plaque active at pH 5–9 (26).
Fig. 2.
Probability density for the solvent-accessible surface area (SAS) of the CHC residues, Leu-17-Ala-21, in Aβ(1–28) and Aβ(10–42) at pH 2 (red), 4 (orange), 6 (green), and 8 (blue).
pH-Dependent β-Turn Formation.
To further understand the link between folding and aggregation in Aβ, we probed the structural features of residues around 24–28. Solution NMR data of Aβ(1–40) and Aβ(1–42) suggested turn-like structures in residues 20–26 (4) or a turn in residues 22–25 (27). Solution NMR data of Aβ(10–35) (24) and Aβ(21–30) (28) indicated a double turn in residues 22–27 and 24–28, respectively. The formation of the turn is intriguing because it coincides with the structural loop region in the fibrils of Aβ(1–40) (20) and Aβ(1–42) (21). The existence and nature of the electrostatic interaction between Lys-28 and either Asp-23 or Glu-22 has been highly debated. The NMR data for Aβ(21–30) revealed long-range electrostatic interactions between Glu-22/Asp-23 and Lys-28 (28). Whereas an intramolecular salt bridge between Asp-23 and Lys-28 was proposed for the fibrils of Aβ(1–40) (20), the same salt bridge was suggested to be intermolecular in the fibrils of Aβ(1–42) (21). The stability of the turn (24–27) in Aβ(10–35) and the turn (24–28) in Aβ(21–30) has previously been tested in simulation studies (29, 30) using the respective NMR structure models (24, 28). The simulation study of Aβ(21–30) also revealed a salt bridge between Glu-22 and Lys-28 (30). In this work, we focus on the role of pH on the turn forming propensity in Aβ(1–28) and Aβ(10–42). Our results are not biased toward any particular sequence location for the turn because we performed folding simulations starting from an extended configuration.
Because our secondary structure analysis shows that residues between 22 and 29 have low helix propensity at all pH but 8 (Fig. 1A), we scanned this sequence for tetrapeptides that form a β-turn according to a widely used definition based on the backbone (φ, ψ) angles (31). At pH 2, the probability for β-turn formation in the region 22–29 of Aβ(1–28) and Aβ(10–42) is <20% (Fig. 3A). In contrast, at pH 4–8, residues 23–26 form a β-turn with a probability of 35–70% in both peptides (Fig. 3A). Considering that the intrinsic turn-forming potential in these resides (AspValGlySer) is independent of pH (32), the significantly increased turn propensity at pH >2 must be due to the interactions of nearby titratable residues. Consequently, we proceeded to examine whether the turn formation is facilitated by the electrostatic attraction between Glu-22 and Lys-28, or between Asp-23 and Lys-28, as was suggested for Aβ(21–30) (28).
Fig. 3.
Correlation between the formation of a β-turn in residues 23–26 and the electrostatic interaction between Glu-22 and Lys-28. (A) Probability density for the formation of a β-turn as a function of pH. (B) Relative free energy, ΔG, as a function of the Glu-22-Lys-28 distance in the presence (solid lines and absence (dashed lines) of the turn from simulations at pH 4. The distance between Glu-22 and Lys-28 is defined as the minimum separation between the carboxylate oxygen atom in Glu-22 and the amino nitrogen atom in Lys-28. ΔG is defined as −RTlnP, where P is the probability density for the distance to fall into one of the 22 bins between 2 and 24 Å.
Analysis of the probability distribution for the distance between Glu-22 and Lys-28, and between Asp-23 and Lys-28, revealed that the electrostatic interaction between Glu-22/Asp-23 is strongest at pH 4 and weakest at pH 8 (SI Fig. 8). At pH 4, the probability density for the distance between Glu-22 and Lys-28 is bimodal, revealing two conformational families in simulations of both Aβ(1–28) and Aβ(10–42). One has a Glu-22… Lys-28 distance <6 Å, whereas the other has a Glu-22… Lys-28 distance >12 Å (SI Fig. 8). In contrast, the probability for the Asp-23… Lys-28 distance is unimodal, displaying a maximum at 6 Å for Aβ(10–42) but at 12 Å for Aβ(1–28). Thus, we decided to focus on the interaction between Glu-22 and Lys-28. Another reason is that, as will be shown later, this interaction is also involved in the disruption of helix formation. We asked whether the β-turn at 23–26 is favored by one of the two conformational states. Fig. 3B shows the computed relative free energy in the presence and absence of the turn formation. Although the turn exists in both conformations, absence of the turn is seen exclusively in the latter conformation. Thus, our data support the hypothesis that the Coulombic electrostatic interaction Glu-22… Lys-28 promotes stability of the β-turn. However, this interaction is clearly not the only contributing factor as seen in Fig. 3B. In fact, we found that, at pH 8, a hydrophobic interaction between Phe-19 and Leu-34, a residue remote in sequence to the turn region 23–26, is correlated with an increased turn propensity (SI Fig. 9). Considering that the β-turn localizes within the bend region that connects two β-strands in the fibril (20), the decreased turn propensity in the monomer Aβ at pH 2 may be another contributing factor for the retarded aggregation under low-pH conditions (5, 6).
The Role of Residue-Specific Interactions.
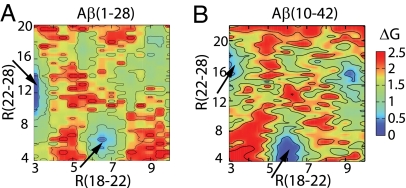
Variation of solution pH allows us to probe the role of residue-specific interactions in modulating the folding of Aβ. A relative free-energy map as a function of the backbone CO… NH distance between Val-18 and Glu-22 and the side-chain distance between Glu-22 and Lys-28 reveals that Glu-22 and Lys-28 are further apart in conformational states that maintain a backbone hydrogen bond between Val-18 and Glu-22 (Fig. 4, upper left arrows). Conversely, the backbone hydrogen bond between Val-18 and Glu-22 is disrupted in states displaying a strong electrostatic interaction between Glu-22 and Lys-28 (Fig. 4, lower right arrows). Thus, the attraction between Glu-22 and Lys-28 not only facilitates the formation of a β-turn as discussed earlier but also destabilizes helix formation in the CHC region at pH 4. This explains the decreased helicity at pH 4 relative to that at pH 2. Although this relationship holds true for pH 6 as well, we found that the nonspecific aromatic interaction between His-14 and Phe-19, which is absent at pH 4 when His-14 is charged, also prevents helix formation in the CHC region (SI Fig. 10). Thus, the helicity of the CHC residues is further reduced as pH is increased from 4 to 6.
Fig. 4.
Correlation between the backbone hydrogen bond formation and the side-chain electrostatic or hydrophobic interactions. Relative free energy, ΔG (in kcal/mol), as a function of the backbone CO… NH distance between Val-18 and Glu-22 (x axis) and the side-chain distance between Glu-22 and Lys-28 (y axis) in the simulation of Aβ(1–28) (A) and Aβ(10–42) (B) at pH 4. The distance between Glu-22 and Lys-28 is defined as the minimum separation between the carboxylate oxygen atom of Glu-22 and the amino nitrogen atom of Lys-28.
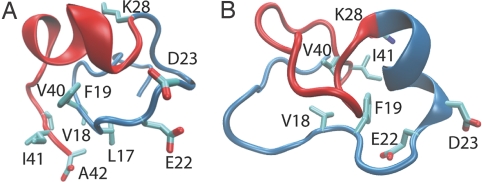
An obvious explanation for the increased solvent exposure in the CHC region as the solution medium becomes less acidic (pH 2–6) is the reduced probability for nascent helix formation. However, why are the CHC residues sequestered from solvent despite their low helix propensity at pH 8? We suggest that this is due to shielding by the C-terminal residues. Representative conformations at pH 8 obtained from clustering reveal hydrophobic interactions between the CHC residues Leu-17, Val-18, and Phe-19 and the C-terminal residues Val-40, Ile-41, and Ala-42, because the CHC region is oriented parallel to the C-terminal segment. This orientation is further stabilized by a backbone β-bridge between Phe-19 and Ile-41 (Fig. 5A). Alternatively, the CHC and C-terminal residues Val-18, Phe-19, Val-40, and Ile-41 can form a hydrophobic cluster in a conformation that favors helix formation at residues 23–28 (Fig. 5B). Although the latter seems to be in conflict with the significant β-turn propensity in residues 23–26, an argument involving the helix dipole may offer a clue. At pH 8, because the electrostatic interaction between Glu-22/Asp-23 and Lys-28 is very weak, the main effect of these charged residues is stabilization of the helix dipole (33), which allows the formation of a nascent helix at 23–28 when these residues do not adopt a β-turn conformation.
Fig. 5.
Representative conformations of Aβ(10–42) at pH 8. (A) Centroid of the largest conformational cluster. This conformation contains a β-turn at residues 23–26 and a parallel orientation of the CHC region with respect to the C-terminal residues. The latter is stabilized by a β-bridge between Phe-19 and Ile-41 as well as the side-chain hydrophobic interactions between the CHC residues Leu-17, Val-18, and Phe-19 and the C-terminal residues Val-40, Ile-41, and Ala-42. (B) Centroid of the second largest conformational cluster. This conformation contains a helix at residues 23–28 as well as a hydrophobic cluster composed of residues Val-18, Phe-19, Val-40, and Ile-41.
Mutations of Glu-22 (Glu-22/Gln and Glu-22/Gly) and Asp-23 (Asp-23/Asn) have been linked to early-onset Alzheimer's disease and higher aggregation rates for Aβ in vitro (34). Our data suggest that the electrostatic interaction between Glu-22 and Lys-28 may accelerate aggregation by reducing nascent helix formation in the CHC region and providing stabilization for a β-turn at 23–26. Our data also shows that both Glu-22 and Asp-23 can interact with Lys-28, whereas the former interaction appears to be slightly stronger. Although we do not have direct evidence for the explanation of the effects due to mutations of Glu-22 and Asp-23, our data are consistent with the proposal that deletion of a charge at either of these residues may strengthen the alternative electrostatic interaction. thereby promoting the aggregation of Aβ (28).
Conclusions
Although our calculated pH profile of the total helix propensity for Aβ(1–28) and Aβ(10–42) is in accord with the pH-dependent helix formation of Aβ peptides in TFE solution as measured by CD (3), other conformational features that emerged from our simulations are consistent with a collapsed coil model consisting of loops, strands, and turns, as proposed based on the solution NMR data of Aβ(10–35) (24, 26). Our simulation data revealed a β-turn in residues 23–26 for both Aβ(1–28) and Aβ(10–42) at a pH >4, consistent with the solution NMR data of the full-length (4, 27) as well as various fragments of Aβ(24, 28). A kink or hinge in this region has also been found in the full-length Aβ in a water–TFE (35) or water–micelle environment (36–38). Thus, our theoretical study combined with the existing experimental data led us to propose that the full-length Aβ is in a conformational equilibrium comprising an ensemble of rapidly interconverting compact coil states with nascent helical fragments, which are transformed into longer helical fragments in the presence of TFE (35, 39) or membrane-mimicking detergent (36–38).
The most striking finding from our simulations is that the helix propensity and solvent exposure of the CHC region as well as the β-turn formation for residues 23–26 in Aβ(1–28) and Aβ(10–42) are strongly modulated by solution pH. At pH 6, the CHC residues display the lowest helix propensity and the highest degree of solvent exposure, all of which are promoting factors for nonspecific intermolecular hydrophobic association of Aβ peptides. Our theoretical prediction of the significant loss of helix propensity in the CHC region as pH increases to 6 is in agreement with the observation by NMR that the helical region 15–24 in Aβ(1–40) starts to unwind at pH >6 in a water–micelle medium (36). Our data suggest that the CHC region is responsible for the pH-dependent α-helix-to-β-sheet transition in TFE solution. Although our prediction of the pH-dependent sequestration of CHC residues has no direct experimental verification to date, it is nonetheless consistent with the following NMR data. Aβ(10–35) and Aβ(1–40) bind to AD plaques in a pH-dependent manner at pH >5; an increase in pH from 2.1 to 5.6 induces a conformational transition (with unknown details) in residues flanking the CHC region (24, 26). Our simulations showed that specific electrostatic interactions, e.g., between Glu-22 and Lys-28, contribute to the destabilization of nascent helices but promote the formation of a β-turn in residues 23–26 as pH is raised from 2 to 4. The latter is consistent with the NMR data (28) as well as a previous simulation study of Aβ(21–30) (30). Thus, we propose that the residual structure of Aβ is strongly modulated by solution pH through changes in intramolecular electrostatic interactions. Remarkably, at pH 6, Aβ peptides adopt conformations that contain “fibril-friendly” elements, poised to form β-sheet-based oligomers via hydrophobic association of the CHC residues (Fig. 6).
Fig. 6.
A proposed mechanism for the pH-dependent aggregation of Aβ. Depicted are representative structures obtained as the centroids of the most populated conformational clusters under the pH conditions of 2, 4, 6, and 8, respectively. The N-terminal residues 1–28 are shown in blue; the C-terminal residues 29–42 are shown in red. In the most aggregation-prone state (pH 6), the side chains of Leu-17, Val-18, Phe-19, Phe-20, and Ala-21 are shown as van der Waals spheres in pink, gray, cyan, purple, and green, respectively.
The idea that pH-dependence of Aβ oligomerization and fibril formation may be due to the pH-dependent intermolecular electrostatic interactions follows naturally from the observation that proteins precipitate most at the isoelectric point where the net charge is zero (40). We argue that, although minimum charge–charge repulsion at the isoelectric point provides an initial driving force for aggregation, the folding landscape of the monomeric Aβ is strongly modulated by pH, leading to an enhanced intermolecular hydrophobic association that stabilizes the β-sheet-based aggregates. According to our data, the optimum pH condition for aggregation is not the isoelectric point, which requires all histidines to be charged. In contrast, His-13 is neutral at pH 6 in our simulations (SI Table 1). This surprising finding is consistent with the experimental data showing that the pH for maximum turbidity of Aβ(1–40) is close to but does not coincide with the pH for maximum fibril formation (40). It is also consistent with the observation of spherical particulates rather than β-sheet-based fibrils at the isoelectric point of several amyloidogenic proteins (41). Thus, we propose that an optimal pH for structured aggregates is near but not matching the isoelectric point at which amorphous aggregates may be more likely to occur.
In light of the emerging experimental evidence revealing the acidic early endosomes with a pH of 6 as the principal generation site for Aβ (7, 8), and dimers and trimers as the major toxic species (9, 10), our findings support a hypothesis that small Aβ oligomers may form intracellularly before being released into the extracellular medium where they may interfere with synaptic activity or act as seeds to accelerate fibril formation (42). Our work represents an early effort toward understanding the environmental effects on protein folding and aggregation. The pH-dependent intramolecular mechanism of Aβ unveiled by our study may underlie the aggregation phenomena of other natively unfolded proteins such as α-synuclein from Parkinson's disease. The theoretical and computational methodology illustrated here is general and can be applied to elucidate various roles of pH in mediating biological processes.
Materials and Methods
We have described the details of the continuous CPHMD method elsewhere (11, 12). Briefly, this method enables molecular dynamics simulations to be performed simultaneously with the titration of ionizable side chains under an external pH condition in the presence of a generalized Born (GB) implicit solvent model. Combined with a state-of-the-art conformational sampling protocol, the REX algorithm (13), benchmark studies demonstrated that REX-CPHMD simulations can offer accurate and robust first-principles prediction for protein pKa values (16) and atomically detailed molecular mechanisms for pH-dependent protein folding (17, 18). It should be noted that our simulations used a recently improved implicit solvent model, GB with a simple switching (GBSW) (14, 15). Its application to protein folding studies has been validated by comparison between simulated and experimental folding behavior for a series of helix- and β-sheet-based peptides (15, 17).
We carried out REX-CPHMD folding simulations for peptides Aβ(1–28) and Aβ(10–42) using the CHARMM molecular dynamics program (version c33a1) (43), in which the CPHMD and GB with a simple switching methods were implemented, and the MMTSB Tool Set for controlling the REX sampling (44). We used the CHARMM22 (45) force field with the dihedral cross-term corrections (CMAP) (46). A cutoff distance of 24 Å was used for both nonbonded and GB calculations. Simulations of Aβ(1–28) were initiated from an ideal helix structure of an uncapped sequence at pH 2, 4, 6, and 8. The side chains of α-amino, Asp, Glu, His, and α-carboxyl residues were allowed to titrate. A REX simulation at one pH used 16 replicas (60-ns each) occupying an exponentially spaced temperature scale between 298 K and 600 K. A conformational exchange was attempted every 2 ps between replicas adjacent in temperature with an acceptance ratio of 38–42%. The REX-CPHMD simulation of Aβ(10–42) used 20 replicas (50-ns each) exchanging at a ratio of 43–50%. The starting structure was an extended sequence with a free C terminus and an acetylated N terminus. Unless otherwise specified, all analyses were based on the coordinates recorded every 10 ps during the last 30 ns of simulation at 298 K. Previous REX and REX-CPHMD folding simulations for peptides of comparable size using the same implicit solvent model and a similar time scale were able to generate conformational equilibria in agreement with experiment (15, 17). The convergence of the current simulations is further demonstrated in SI Fig. 7.
The secondary structure assignment was performed by using the DSSP program (47). The residue-based helix and turn propensities were obtained as the fractional occurrence of these states during the last 30 ns of each simulation. The total helix content was computed as the fraction of helical residues. This value cannot be directly compared with the helix content deduced from a CD experiment, because the latter depends not only on the number of helical residues but also on the helical length as well as the deviation of backbone torsion angles from the ideal helix values (48). To facilitate comparison with experiment, we derived the helix content Fhelix from a calculated mean residue ellipticity at 222 nm, [θ]calc, using the following relationship (49):
where [θ]coil is the mean residue ellipticity for a complete random coil, taken as 640 (50), whereas [θ]helix is the mean residue ellipticity for a complete helix given as
where Nr is the total number of residues in the peptide. The calculated mean residue ellipticity was obtained as (48)h:
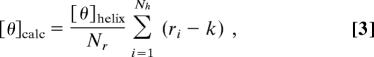 |
where Nh is the number of helical fragments, ri is the number of helical residues according to the DSSP assignment (47), k is the minimum number of helical residues required to produce a CD signal, taken as 3 (49). The unit of mean residue ellipticity is deg·cm2·dmol−1.
The solvent-accessible surface area for the CHC (17–21) and the C-terminal hydrophobic (29–42) residues was calculated by using the Lee and Richard's algorithm (51) with van der Waals radii for all atoms and a solvent probe radius of 1.4 Å.
Conformational clustering of the recorded coordinates was performed by using the GROMACS molecular dynamics software package (52) with a hierarchical algorithm based on the root mean square deviation between backbone atoms with a cutoff of 1 Å (53).
Supplementary Material
Acknowledgments
We thank Drs. Dennis Selkoe, Dominic Walsh, Michael Zagroski, and David Teplow for helpful comments. This work was supported by National Institutes of Health Grants GM57513, GM48807, and RR12255.
Abbreviations
- Aβ
β-amyloid
- AD
Alzheimer's disease
- CHC
central hydrophobic cluster
- CPHMD
constant pH molecular dynamics
- REX
replica exchange
- CD
circular dichroism.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0703832104/DC1.
References
- 1.Dobson CM. Nature. 2003;426:884–890. doi: 10.1038/nature02261. [DOI] [PubMed] [Google Scholar]
- 2.Dyson HJ, Rance M, Houghten RA, Wright PE, Lerner RA. J Mol Biol. 1988;201:201–217. doi: 10.1016/0022-2836(88)90447-0. [DOI] [PubMed] [Google Scholar]
- 3.Barrow CJ, Yasuda A, Kenny PTM, Zagorski MG. J Mol Biol. 1992;225:1075–1093. doi: 10.1016/0022-2836(92)90106-t. [DOI] [PubMed] [Google Scholar]
- 4.Hou L, Shao H, Zhang Y, Li H, Menon NK, Neuhaus EB, Brewer JM, Byeon I-JL, Ray DG, Vitek MP, et al. J Am Chem Soc. 2004;126:1992–2005. doi: 10.1021/ja036813f. [DOI] [PubMed] [Google Scholar]
- 5.Fraser PE, Nguyen JT, Surewicz WK, Kirschner DA. Biophys J. 1991;60:1190–1201. doi: 10.1016/S0006-3495(91)82154-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kirkitadze MD, Condron MM, Teplow DB. J Mol Biol. 2001;312:1103–1119. doi: 10.1006/jmbi.2001.4970. [DOI] [PubMed] [Google Scholar]
- 7.Kaether C, Schmitt S, Willem M, Haass C. Traffic. 2006;7:408–415. doi: 10.1111/j.1600-0854.2006.00396.x. [DOI] [PubMed] [Google Scholar]
- 8.Rajendran L, Honsho M, Zahn TR, Keller P, Geiger KD, Verkade P, Simons K. Proc Natl Acad Sci USA. 2006;103:11172–11177. doi: 10.1073/pnas.0603838103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cleary JP, Walsh DM, Hofmeister JJ, Shankar GM, Kuskowski MA, Selkoe DJ, Ashe KH. Nat Neurosci. 2004;8:79–84. doi: 10.1038/nn1372. [DOI] [PubMed] [Google Scholar]
- 10.Klyubin I, Walsh DM, Lemere CA, Cullen WK, Shankar GM, Betts V, Spooner ET, Jiang L, Anwyl R, Selkoe DJ, Rowan MJ. Nat Med. 2005;11:556–561. doi: 10.1038/nm1234. [DOI] [PubMed] [Google Scholar]
- 11.Lee MS, Salsbury FR, Jr, Brooks CL., III Proteins. 2004;56:738–752. doi: 10.1002/prot.20128. [DOI] [PubMed] [Google Scholar]
- 12.Khandogin J, Brooks CL., III Biophys J. 2005;89:141–157. doi: 10.1529/biophysj.105.061341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sugita Y, Okamoto Y. Chem Phys Lett. 1999;314:141–151. [Google Scholar]
- 14.Im W, Lee MS, Brooks CL., III J Comput Chem. 2003;24:1691–1702. doi: 10.1002/jcc.10321. [DOI] [PubMed] [Google Scholar]
- 15.Chen J, Im W, Brooks CL., III J Am Chem Soc. 2006;128:3728–3736. doi: 10.1021/ja057216r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khandogin J, Brooks CL., III Biochemistry. 2006;45:9363–9373. doi: 10.1021/bi060706r. [DOI] [PubMed] [Google Scholar]
- 17.Khandogin J, Chen J, Brooks CL., III Proc Natl Acad Sci USA. 2006;103:18546–18550. doi: 10.1073/pnas.0605216103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khandogin J, Raleigh DP, Brooks CL., III J Am Chem Soc. 2007;129:3056–3057. doi: 10.1021/ja0688880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Balbach JJ, Ishii Y, Antzutkin ON, Leapman RD, Rizzo NW, Dyda F, Reed J, Tycko R. Biochemistry. 2000;39:13748–13759. doi: 10.1021/bi0011330. [DOI] [PubMed] [Google Scholar]
- 20.Petkova AT, Ishii Y, Balbach JJ, Antzutkin ON, Leapman RD, Delaglio F, Tycko R. Proc Natl Acad Sci USA. 2002;99:16742–16747. doi: 10.1073/pnas.262663499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lührs T, Ritter C, Adrian M, Riek-Loher D, Bohrmann B, Döbeli H, Schubert D, Riek R. Proc Natl Acad Sci USA. 2005;102:17342–17347. doi: 10.1073/pnas.0506723102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kallberg Y, Gustafsson M, Persson B, Thyberg J, Johansson J. J Biol Chem. 2001;276:12945–12950. doi: 10.1074/jbc.M010402200. [DOI] [PubMed] [Google Scholar]
- 23.Tjernberg LO, Näslund J, Lindqvist F, Johansson J, Karlstromi AR, Thyberg J, Terenius L, Nordstedt C. J Biol Chem. 1996;271:8545–8548. doi: 10.1074/jbc.271.15.8545. [DOI] [PubMed] [Google Scholar]
- 24.Zhang S, Iwata K, Lachenmann MJ, Peng JW, Li S, Stimson ER, Lu Y, Felix AM, Maggio JE, Lee JP. J Struct Biol. 2000;130:130–141. doi: 10.1006/jsbi.2000.4288. [DOI] [PubMed] [Google Scholar]
- 25.Chen Y-R, Glabe CG. J Biol Chem. 2006;281:24414–24422. doi: 10.1074/jbc.M602363200. [DOI] [PubMed] [Google Scholar]
- 26.Lee JP, Stimson ER, Ghilardi JR, Mantyh PW, Lu Y-A, Felix AM, Llanos W, Behbin A, Cummings M, Crielunge MV, et al. Biochemistry. 1995;34:5101–5200. doi: 10.1021/bi00015a033. [DOI] [PubMed] [Google Scholar]
- 27.Laurents DV, Gorman PM, Guo M, Rico M, Chakrabartty A, Bruix M. J Biol Chem. 2005;280:3675–3685. doi: 10.1074/jbc.M409507200. [DOI] [PubMed] [Google Scholar]
- 28.Lazo ND, Grant MA, Condron MC, Rigby AC, Teplow DB. Protein Sci. 2005;14:1581–1596. doi: 10.1110/ps.041292205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Massi F, Peng JW, Lee JP, Straub JE. Biophys J. 2001;80:31–44. doi: 10.1016/S0006-3495(01)75993-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baumketner A, Bernstein SL, Wyttenbach T, Lazo ND, Teplow DB, Bowers MT, Shea J-E. Protein Sci. 2006;15:1239–1247. doi: 10.1110/ps.062076806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilmot CM, Thornton JM. Protein Eng. 1990;3:479–493. doi: 10.1093/protein/3.6.479. [DOI] [PubMed] [Google Scholar]
- 32.Hutchinson EG, Thornton JM. Protein Sci. 1994;3:2207–2216. doi: 10.1002/pro.5560031206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shoemaker KR, Kim PS, York EJ, Stewart JM, Baldwin RL. Nature. 1987;326:563–567. doi: 10.1038/326563a0. [DOI] [PubMed] [Google Scholar]
- 34.Pawar AP, DuBay KF, Zurdo J, Chiti F, Vendruscolo M, Dobson CM. J Mol Biol. 2005;350:379–392. doi: 10.1016/j.jmb.2005.04.016. [DOI] [PubMed] [Google Scholar]
- 35.Sticht H, Bayer P, Willbold D, Dames S, Hilbich C, Beyreuther K, Frank RW, Rösch P. Eur J Biochem. 1995;233:293–298. doi: 10.1111/j.1432-1033.1995.293_1.x. [DOI] [PubMed] [Google Scholar]
- 36.Coles M, Bicknell W, Watson AA, Fairlie DP, Craik DJ. Biochemistry. 1998;37:11064–11077. doi: 10.1021/bi972979f. [DOI] [PubMed] [Google Scholar]
- 37.Shao H, Chuan Jao S, Ma K, Zagorski MG. J Mol Biol. 1999;285:755–773. doi: 10.1006/jmbi.1998.2348. [DOI] [PubMed] [Google Scholar]
- 38.Crescenzi O, Tomaselli S, Guerrini R, Salvadori S, D'Ursi AM, Temussi PA, Picone D. Eur J Biochem. 2002;269:5642–5648. doi: 10.1046/j.1432-1033.2002.03271.x. [DOI] [PubMed] [Google Scholar]
- 39.Barrow C, Zagorski MG. Science. 1991;253:179–183. doi: 10.1126/science.1853202. [DOI] [PubMed] [Google Scholar]
- 40.Guo M, Gorman PM, Ricob M, Chakrabartty A, Laurents DV. FEBS Lett. 2005;579:3574–3578. doi: 10.1016/j.febslet.2005.05.036. [DOI] [PubMed] [Google Scholar]
- 41.Krebs MRH, Devlin GL, Donald AM. Biophys J. 2007;92:1336–1342. doi: 10.1529/biophysj.106.094342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Selkoe DJ. Nat Cell Biol. 2004;6:1054–1061. doi: 10.1038/ncb1104-1054. [DOI] [PubMed] [Google Scholar]
- 43.Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. J Comput Chem. 1983;4:187–217. [Google Scholar]
- 44.Feig M, Karanicolas J, Brooks CL., III J Mol Graphics Model. 2004;22:377–395. doi: 10.1016/j.jmgm.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 45.MacKerell AD, Jr, Bashford D, Bellott M, Dunbrack RL, Jr, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, et al. J Phys Chem B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 46.Feig M., MacKerell AD, Jr, Brooks CL., III J Phys Chem B. 2003;107:2831–2836. [Google Scholar]
- 47.Kabsch W, Sander C. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 48.Hirst J, Brooks CL., III J Mol Biol. 1994;243:173–178. doi: 10.1006/jmbi.1994.1644. [DOI] [PubMed] [Google Scholar]
- 49.Myers JK, Pace CN, Scholtz JM. Proc Natl Acad Sci USA. 1997;94:2833–2837. doi: 10.1073/pnas.94.7.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rohl CA, Chakrabartty A, Baldwin RL. Protein Sci. 1996;5:2623–2637. doi: 10.1002/pro.5560051225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee B, Richards FM. J Mol Biol. 1971;55:379–380. doi: 10.1016/0022-2836(71)90324-x. [DOI] [PubMed] [Google Scholar]
- 52.Lindahl E, Hess B, van der Spoel D. J Mol Model. 2001;7:306–317. [Google Scholar]
- 53.Daura X, Gademann K, Jaun B, Seebach D, van Gunsteren WF, Mark AE. Angew Chem Int Ed. 1999;38:236–240. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.