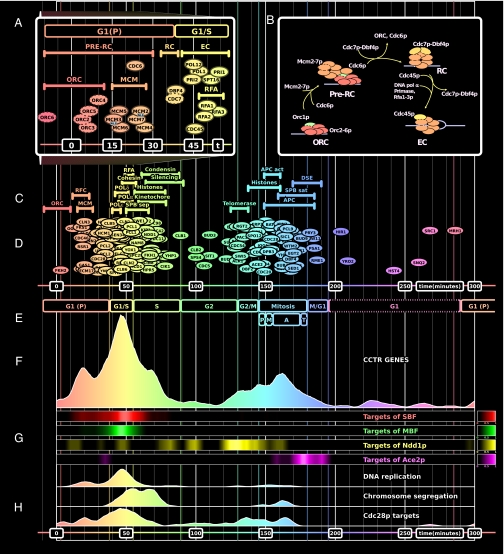
Fig. 4.
Transcriptional program of the yeast cell cycle. (A and B) Proteins involved in DNA replication initiation, color-coded according to timing of their expression. The prereplication complex (pre-RC) undergoes several changes before an elongation complex (EC), capable of initiating DNA synthesis, is formed (24). The order of expression (A) agrees with the order in which gene products are needed (B). In A, dotted outlines denote non-CCTR genes. Note the two groups of MCM subunits, each containing one nuclear localization signal (NLS). In B, solid outlines denote primary expression peak; dashed outlines denote secondary (lower scoring) peak. The only exception to just-in-time transcription is ORC1. (C) Timing of CCTR complexes. DSE, daughter-cell-specific expression program; APC act, APC activation; SPB sat, spindle pole body satellite formation; SPB sep, spindle pole body duplication and separation. (D) Peaks of selected CCTR genes. (E) Phases and subphases of the cell cycle. Note new prereplicative G1 (P) phase. (F) Histogram of expression peaks of CCTR genes. (G) Peaks of transcripts regulated by selected cell cycle transcription factors (11, 20, 21, 29, 30). Note the differences in expression of MBF and SBF targets. (H) Histograms of peaks of CCTR genes involved in selected cell cycle functions. Compare peaks of predicted Cdc28p targets (26) vs. peaks of CDC28 (D).