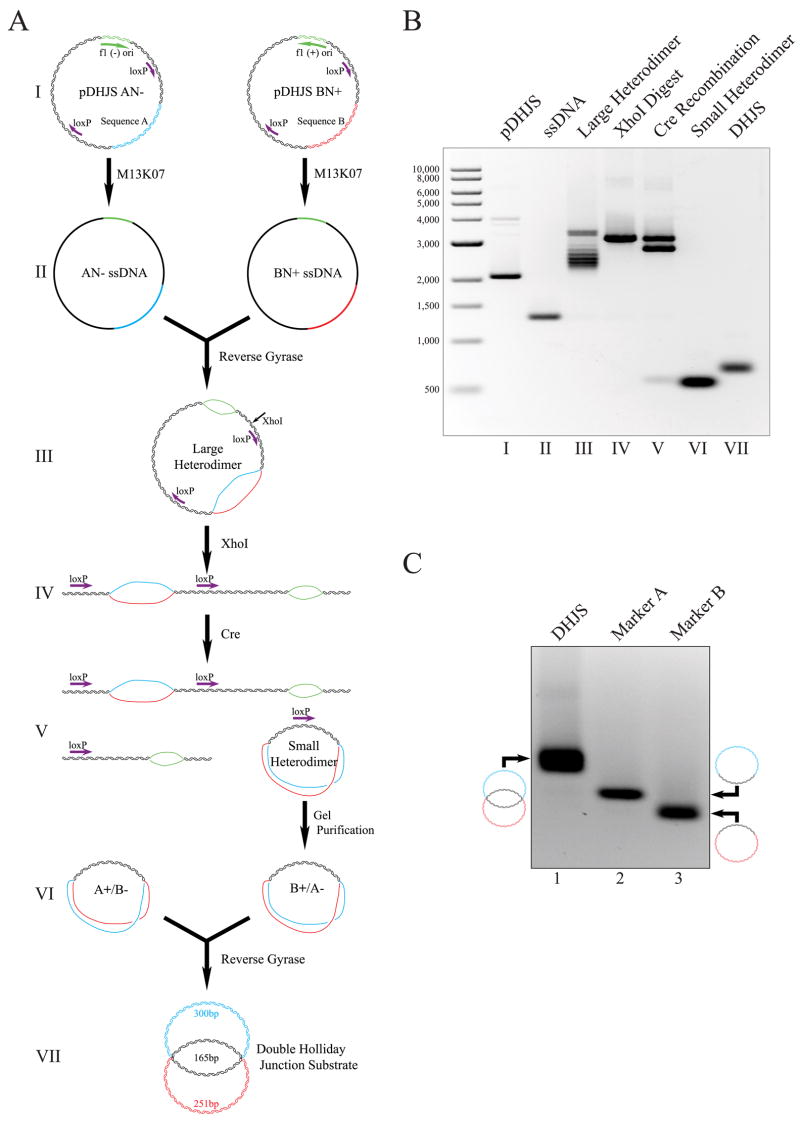
FIGURE 2. Synthesis of a topologically constrained double Holliday Junction substrate.
A, schematic of synthesis. The substrate is derived from four phagemid vectors, containing tandem loxP sites and differing with either sequence A or B, and with the f1 origin in both the plus and minus orientations for each. For simplicity, reactions for only two of the plasmids are shown (I). Using the helper phage M13KO7, ssDNA circles are expressed, purified (II), mixed, and linked using reverse gyrase to yield a large heterodimer intermediate (III). The large heterodimer is linearized by digestion with XhoI (IV), and the small heterodimer was excised and circularized utilizing Cre recombinase and the loxP sites flanking the A/B bubble (V ). The small heterodimer is gel-purified away from the other reaction products of the Cre reaction and is mixed with the complementary small heterodimer created from the other two plasmids in a parallel reaction (VI). The small heterodimers are linked together with reverse gyrase to yield the double Holliday junction substrate (VII). This illustration is not drawn to scale. B, agarose gel electrophoresis of intermediates of DHJS synthesis. The first lane contains a size marker, with the size of the bands denoted to the left of the gel in base pairs. The roman numerals below the lanes correspond to the stages of synthesis in A. C, agarose gel electrophoresis of the marker molecules. Lane 1 contains the DHJS; lane 2, Marker A; lane 3, Marker B. Diagrams of the structure of these dsDNA molecules are shown to the left and right of the gel.