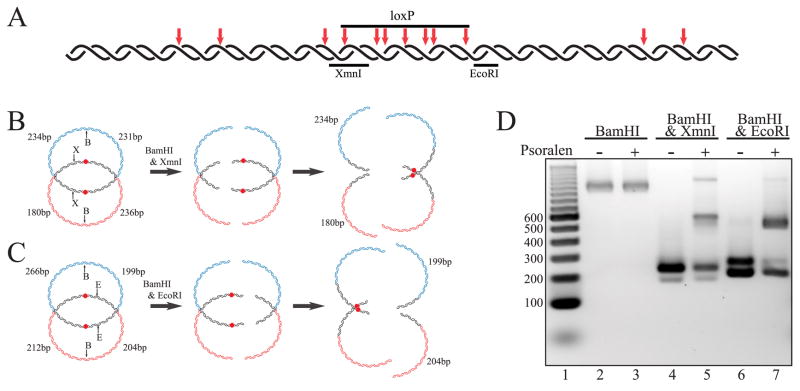
FIGURE 4. Digestion of psoralen cross-linked DHJS is consistent with the predicted structure.
A, the predicted cross-linking sites of psoralen within the homologous region. Psoralen preferentially binds and cross-links at TA sequences, with the repeated TA sequence being particularly good sites. TA dinucleotides are indicated with red arrows, with double arrows showing the location of the TATA sequences in the loxP site. B, the predicted outcomes of BamHI and XmnI digestion of the cross-linked DHJS. Psoralen cross-links are represented by red dots in the center of the homologous region. The XmnI, BamHI double digest should produce 236-, 234-, 231-, and 180-bp bands if the HJs are left unconstrained. However, if the DHJS is cross-linked in the loxP sequence, then this double digest will show a diminished ~235-bp heterogeneous band with the concomitant appearance of a slower migrating species. C, the predicted outcomes of BamHI and EcoRI digestion of the cross-linked DHJS. The EcoRI, BamHI double digest should produce 266-, 212-, 204-, and 199-bp bands if the HJs are left unconstrained. However, if the DHJS is cross-linked in the loxP sequence, then this double digest will show a nearly eliminated 266-bp band and diminished ~200-bp heterogeneous band, with the appearance of a slower migrating species. D, double digestion of the cross-linked DHJS shows the predicted pattern. Lane 1 contains a DNA size marker, with the sizes of the relevant bands denoted to the left of the gel in base pairs. Digestion of the DHJS by BamHI is not effected by the cross-linking (compare lane 3 with lane 2). The XmnI, BamHI double digest of the cross-linked DHJS shows a shifted banding pattern consistent with cross-linking within the loxP sequence (compare lane 4 with lane 5). The EcoRI, BamHI double digest of the cross-linked DHJS also shows a shifted banding pattern consistent with cross-linking within the loxP sequence (compare lane 6 with lane 7).