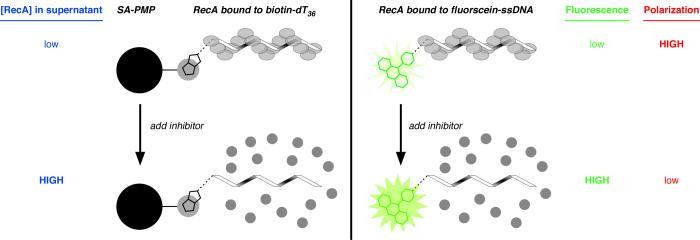
Fig. 2.
Cartoons depicting three assays to assess the extent of assembly of a RecA-DNA filament. In all variations of the assay, an oligonucleotide (32 to 36 nts in length) is incubated with RecA to allow filament assembly, then a putative inhibitor of filament assembly is added. (Left) Assay for RecA assembly on biotin-dT36 associated with streptavidin-coated paramagnetic particles (SA-PMP). If filament assembly on biotin-dT36 is disrupted, any RecA in the supernatant can be detected direclty by colorometric protein quantification after the RecA bound to biotin-dT36 is pulled down using a magnet (blue text). (Right) Assay for RecA assembly on fluorescein-ssDNA using fluorescein emission (either total emission or fluorescence polarization) as the observable parameter. If filament assembly on a 32-nt ssDNA conjugated to fluorescein at the 5′ end is disrupted, the RecA-bound state emits a low total fluorescence signal (green text) and high fluorescence polarization signal (red text); conversely, the RecA-free oligonucleotide state is characterized by high total emission and low fluorescence polarization.