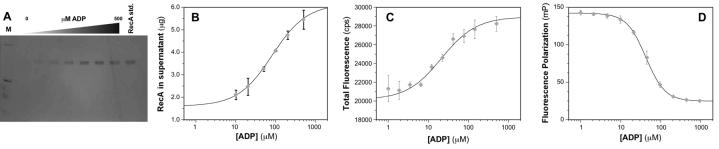
Fig. 3.
Dependence of extent of RecA-DNA filament assembly on concentration of added ADP. (A) Silver-stained gel from SDS-PAGE fractionation of the RecA-DNA filament assembly reactions (Fig. 2, left) in the presence of increasing amounts of ADP. The lane labeled “RecA std.” contains 1.25 μg RecA protein, the maximum amount of protein that can be released in the assay. The concentration of ADP varied between 0 to 500 μM. (B) Plot of amount of unbound RecA, determined using Bradford assay for protein in the supernatant, as a function of ADP concentration for the RecA-DNA filament assembly assay. The data points represent the mean ± one standard deviation of at least three independent experiments, which were identical to those used to create (A). The smooth curve represents the best-fit binding isotherm (Kd = 80 ± 30 μM) as described in Materials and Methods. (C) Plot of total fluorescence in counts per second (cps) as a function of ADP concentration for the RecA-DNA filament assembly assay (Fig. 2, right). The data points represent the mean ± one standard deviation of at least three independent experiments. The smooth curve represents the best-fit binding isotherm (Kd = 20 ± 10 μM) as described in Materials and Methods. (D) Plot of fluorescence polarization in millipolarization units (mP) as a function of ADP concentration for the RecA-DNA filament assembly assay (Fig. 2, right). The data points represent the mean ± one standard deviation of at least three independent experiments. The smooth curve represents the best-fit binding isotherm (Kd = 42 ± 3 μM) as described in Materials and Methods.