Figure 4.
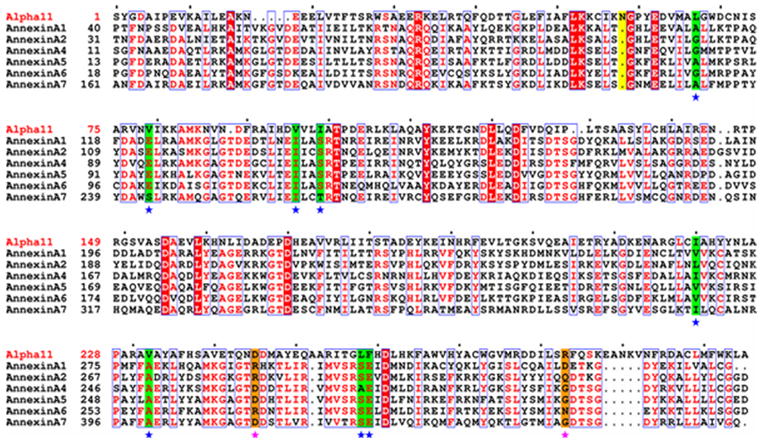
Sequence alignment of alpha-11 giardin and annexins A1, A2, A4, A5, A6 and A7. Note that the first amino acid is a serine residue in the alpha-11 giardin sequence since the first methionine is absent due to the presence of an N-terminal glutathione-S-transferase (GST) tag that was introduced during cloning. Residues denoted as white letters on red background are strictly conserved. Residues boxed in blue and denoted in red are homologous, while residues denoted in black are non-homologous in this group. Highlighted in green and indicated below with a blue star are the residues that line the hydrophobic core located between repeats I/IV and II/III. Highlighted in orange and indicated below with a pink star are residues Asp245 and Arg283 (alpha-11) that form a salt bridge in the AB loop of repeat IV preventing a calcium ion from binding and homologues of these alpha-11 residues in the annexin sequences are highlighted in orange. Highlighted in yellow is Asn58 of alpha-11 giardin which is involved in the type IIIb calcium coordinating site in the DE loop of repeat I. The sequence alignment was performed using CLUSTALW45,46 and the figure was prepared with the program ESPript47.
