Abstract
During gastrulation in Drosophila, ventral cells change shape, undergoing synchronous apical constriction, to create the ventral furrow (VF). This process is affected in mutant embryos lacking zygotic function of the folded gastrulation (fog) gene, which encodes a putative secreted protein. Fog is an essential autocrine signal that induces cytoskeletal changes in invaginating VF cells. Here we show that Fog is also required for nervous system development. Fog is expressed by longitudinal glia in the central nervous system (CNS), and reducing its expression in glia causes defects in process extension and axon ensheathment. Glial Fog overexpression produces a disorganized glial lattice. Fog has a distinct set of functions in CNS neurons. Our data show that reduction or overexpression of Fog in these neurons produces axon guidance phenotypes. Interestingly, these phenotypes closely resemble those seen in embryos with altered expression of the receptor tyrosine phosphatase PTP52F. We conducted epistasis experiments to define the genetic relationships between Fog and PTP52F, and the results suggest that PTP52F is a downstream component of the Fog signaling pathway in CNS neurons. We also found that Ptp52F mutants have early VF phenotypes like those seen in fog mutants.
Keywords: Drosophila nervous system development, receptor tyrosine phosphatase, glia, motor axon guidance, epistasis, ventral furrow development, autocrine signaling, Folded Gastrulation
Introduction
During gastrulation in Drosophila, two groups of cells invaginate and form the primordia of the mesoderm and endoderm. Cells located within a strip around the ventral midline change shape, undergoing synchronous apical constriction, to create the ventral furrow (VF). At the posterior end, apical constriction of a circular group of cells forms the posterior midgut (PMG) invagination, which encloses the pole cells (Sweeton et al., 1991; Costa et al., 1993).
The mechanisms involved in the induction of cell shape changes during gastrulation are not well understood. However, several mutations have been identified that affect these processes. Embryos lacking zygotic function of the folded gastrulation (fog) gene undergo asynchronous changes in cell shape, resulting in an irregular VF, and do not form the PMG invagination [Costa et al., 1994]. Identical phenotypes are observed in embryos lacking maternally contributed concertina (cta) gene function (Parks and Weischaus., 1991).
fog encodes a large protein with an N-terminal hydrophobic region that resembles a signal sequence, suggesting that at least part of it is secreted. It is expressed by the cells that will invaginate to form the VF and PMG (Costa et al., 1994). Cta is a G protein alpha subunit, and it is uniformly distributed within the blastoderm embryo (Parks and Wieschaus., 1991). The similarities in the loss-of-function (LOF) and gain-of-function (GOF) phenotypes of fog and cta, and the observation that cta is epistatic to fog, have led to the proposal that Fog is a ligand for an as yet unidentified G protein-coupled receptor (GPCR) that couples to Cta (Morize et al., 1998).
The mechanisms involved in Fog signaling have been examined primarily in the context of gastrulation. It has been shown that Fog is an autocrine signal that induces cytoskeletal changes in invaginating VF cells (Dawes-Hoang et al., 2005). Exposure to the Fog signal causes translocation of nonmuscle myosin from the basal to the apical cell surface, and this facilitates apical constriction of VF cells (Dawes-Hoang et al., 2005). Fog is also required for development of the salivary glands and Malphigian tubules (Lammel and Saumweber, 2000).
Our laboratory has focused on examining the functions of receptor tyrosine phosphatases (RPTPs) in regulation of CNS and motor axon guidance during Drosophila embryogenesis. Like the other RPTPs, PTP52F is expressed primarily in the embryonic CNS. However, in early embryos, expression of the protein is also observed in cells surrounding the VF (Schindelholz et al., 2001). More recently, we discovered that Ptp52F mutants have fog-like VF phenotypes (Supp. Fig. 2; A. Schmid, B. Schindelholz, and K.Z., unpublished). The similarity in VF phenotype between fog and Ptp52F led us to explore the possibility of an interaction between these two genes in the embryonic CNS.
In this paper, we characterize the expression of Fog in the embryonic CNS. We show that Fog is expressed in glia and neurons, and has distinct roles in the two cell types. Fog expressed by longitudinal glia is required for glial ensheathment of CNS axons. Neuronal Fog regulates axon guidance, and Fog signaling in neurons requires the PTP52F receptor.
Materials and Methods
Fly Stocks
fog4a6 and fog4a6, hkb::fog flies were obtained from the laboratories of E. Wieschaus and M Leptin, respectively. EP52F, an EP insertion upstream of the Ptp52F gene, was obtained from Florenci Serras. Ptp52F18.3, a null allele of Ptp52F (Schindelholz et al., 2001) was used for the epistasis experiment of Fig. 5. For the heat shock experiment of Fig. 1, a hs-GAL4, UAS-GFP line was used. C155-GAL4, elav-GAL4 and Repo-GAL4 were obtained from the Bloomington stock centre. Htl-GAL4 was obtained from Alicia Hidalgo.
Figure 5. Fog and PTP52F CNS overexpression phenotypes, and suppression of the Fog overexpression phenotype by loss of Ptp52F function.
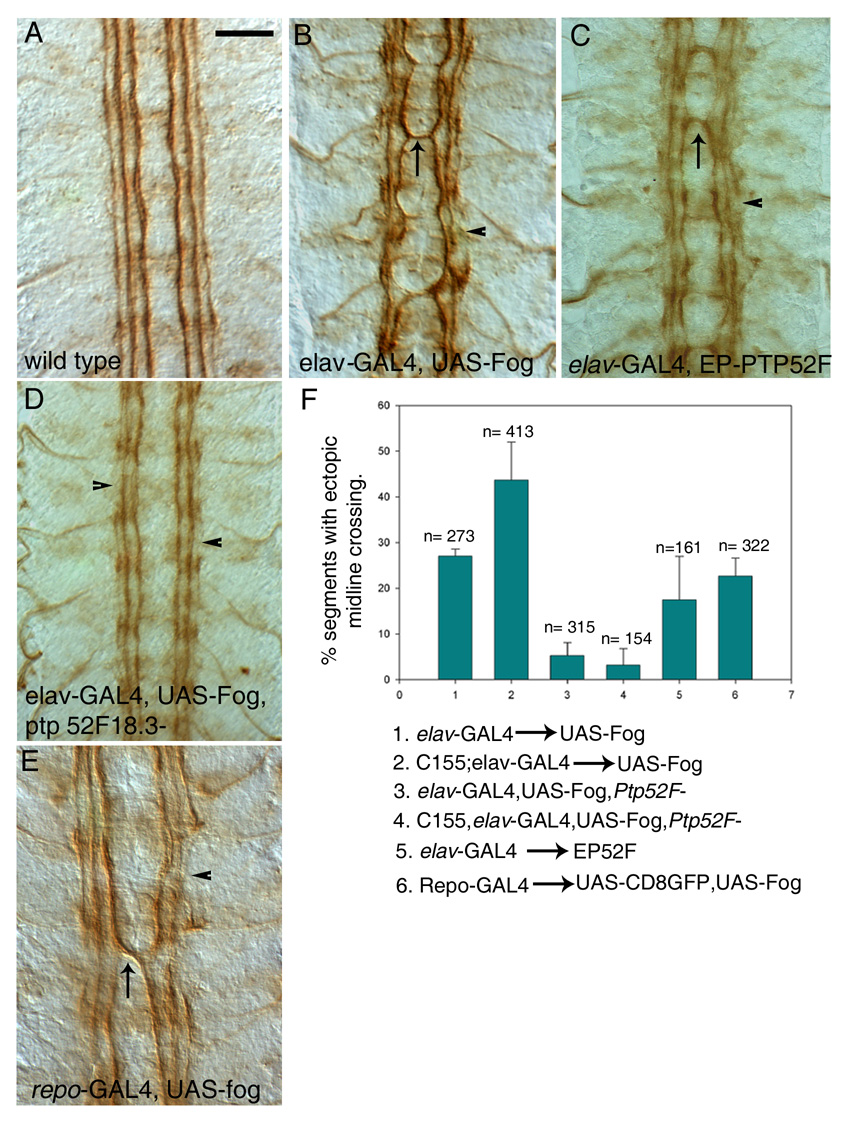
Late stage 16 embryos were stained with mAb 1D4 (brown) and dissected. Four segments are shown; anterior is up.
A) Wild-type (Oregon R) embryo. Note that the three 1D4-positive longitudinal axon bundles do not cross the midline.
(B) Elav-GAL4::UAS-fog. All three longitudinal bundles are disorganized, and often fused to each other. Breaks in the bundles are often present, especially in the outer bundle (arrowhead). Frequent ectopic midline crossing by the inner bundle is observed (arrow).
(C) Elav-GAL4::EP52F. A phenotype similar to (B) is observed, with occasional bundle fusions and breaks (arrowhead), and ectopic midline crossing (arrow).
(D) Elav-GAL4::UAS-fog, Ptp52F18.3/Ptp52F18.3. Removal of Ptp52F function in Fog overexpression embryos suppresses ectopic midline crossing.
(E) Repo-GAL4::UAS-fog. Ectopic midline crossing is also seen when Fog is overexpressed in glia (arrow). Breaks are frequently observed in the outer longitudinal bundles (arrowhead).
(F) Bar graph showing percentages of segments with ectopic midline crossing observed in different genotypes. The numbers above the bars indicate the number of segments scored for each genotype. Scale bar in (A), 10 µm (applies to all panels).
Figure 1. Expression of fog mRNA and protein in dissected stage 16 embryos.
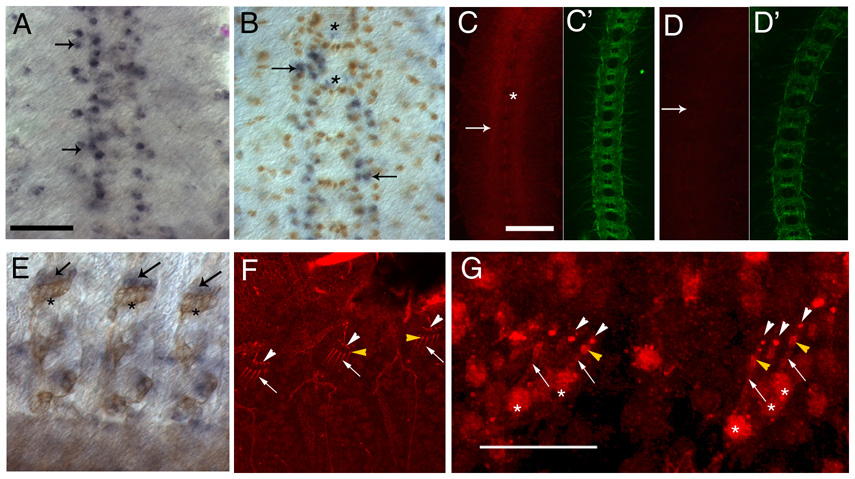
(A and B) fog mRNA expression in the CNS, detected by in situ hybridization using alkaline phosphatase (AP) histochemistry. The CNS (anterior up) is vertically oriented at the center of the panels; four segments are shown. (A) fog mRNA (blue) is expressed in a subset of CNS cells (arrows). Note that the pattern is not precisely bilaterally symmetric and is not entirely reproducible between segments. (B) fog in situ hybridization co-stained with anti-Repo antibody (brown). Most fog positive cells also express Repo (arrows), but some are Repo-negative (asterisks).
(C and D) The CNS in a wild-type (C and C’) and a hs-GAL4::UAS-dsfogRNA (fog RNAi) (D and D’) embryo stained with anti-Fog antibody(red) and mAbBP102 (green), which is a marker for CNS axons. Fog is expressed at highest levels in the CNS axon ladder (C, arrow). Midline glia are indicated by an asterisk in C. Axonal staining is reduced in fog RNAi embryos (D, arrow).
(E) fog mRNA expression (blue, arrow) in peripheral chordotonal organs (also stained with mAb22C10 (brown), which is a marker for peripheral sensory neurons (asterisks indicate chordotonal neuron cell bodies)). fog expression is observed in the region of the scolopale and cap cells (arrows). Anterior is to the left in (E–G).
(F) Expression of Fog protein (red) in chordotonal organs, detected by immunofluorescence. Protein is observed in the dendrites of the sensory neurons (arrows), scolopale cells (bulb-like shapes at the end of the dendrites, yellow arrowheads), and cap cells (white arrowheads).
(G) Chordotonal organs stained with anti-Fog (red) in the absence of detergent. Expression in the dendritic shaft of the sensory neuron (arrows), the scolopales (yellow arrowheads), and the cap cells (white arrowheads) can be detected, indicating that Fog is present at the cell surface. The asterisks mark the cell bodies of the sensory neurons. Note that background staining is higher when this method is used. Scale bar for panels A and B is in panel A and corresponds to 10 µm, for C–F it is in panel C and corresponds to 10 µm, and the scale bar in G corresponds to 25µm.
Generation of transgenic lines
The 1.6 kb full length Fog cDNA was cloned into the pUAST vector and the plasmid DNA was injected into 0–1hr embryos using standard methods. Genetic crosses were carried out to establish stable transgenic lines
To obtain UAS-fog dsRNA transgenic lines, an inverted repeat sequence of the fog cDNA corresponding to amino acids 151–421 was first generated as follows: using two independent 5’ primers and a common 3’ primer, two kinds of PCR products were generated. The 5’ primers were designed such that they carried either a XhoI or a XbaI site at the 5’ end. The common 3’ primer was designed with a BamHI site followed by an EcoRI site at the 5’ end. The first PCR product was cloned into pBluescript using the XbaI and EcoRI sites; the second product was cloned in the opposite orientation using the BamHI present in the first PCR product and the XhoI site in pBluescript. The resulting inverted repeat sequence was then subcloned into pUAST using the XhoI and XbaI sites and transformed using Stbl-2 cells (Invitrogen). The pUAST construct was then injected into 0–1 hr embryos using standard methods to obtain transgenic lines. Several such lines were analyzed, and all had equivalent phenotypes. The neuronal and glial RNAi experiments were carried out at room temperature (23°C) and 29°C, respectively.
in situ hybridization and immunohistochemistry
in situ hybridization was carried out using sense and anti-sense digoxigenin labeled RNA as probes. The hybridization and detection procedures used have been previously described in (Kraut et al., 2001). No signal was detected with the sense probe.
Immunohistochemical analysis was carried out on embryos collected from crosses between flies of the appropriate genotypes. Embryos of the desired genotype were identified by the absence of staining of with rabbit anti beta-galactosidase (Invitrogen), which detects the presence of the lacZ gene on the balancer chromosome. The following antibodies were used : anti-Fog raised against the full length protein (1:100 gift of N. Fuse), anti-Fog (N-terminal antibody) raised against th first 300 amino acids (1:50, gift of the Wieschaus lab), chicken anti-GFP (1:500, Chemicon), anti- Twist (1:5000, gift of S.Roth). Monoclonal antibodies mAb 1D4, mAb 22C10 and anti-Repo (Developmental Studies Hybridoma Bank, Iowa, United States) were used at 1:10, 1:50 and 1:50 respectively. Embryo fixation and immunohistochemistry using horseradish preoxidase and fluorescence was performed as described in (Patel, 1994). All embryos stained using horseradish peroxidase were photographed with a Magnafire digital camera on a Zeiss Axioplan microscope using Nomarski optics.
For the side view of the CNS in Fig 2, dissected CNS of stained embryos were mounted on their side and images were taken as a z-series. A projected image from the z-series was obtained using the Zeiss confocal software. Subsequent processing was done using Adobe Photoshop7.0.
Figure 2. Glial morphology and organization are altered in fog glial RNAi, fog mutant, and Fog overexpression embryos.
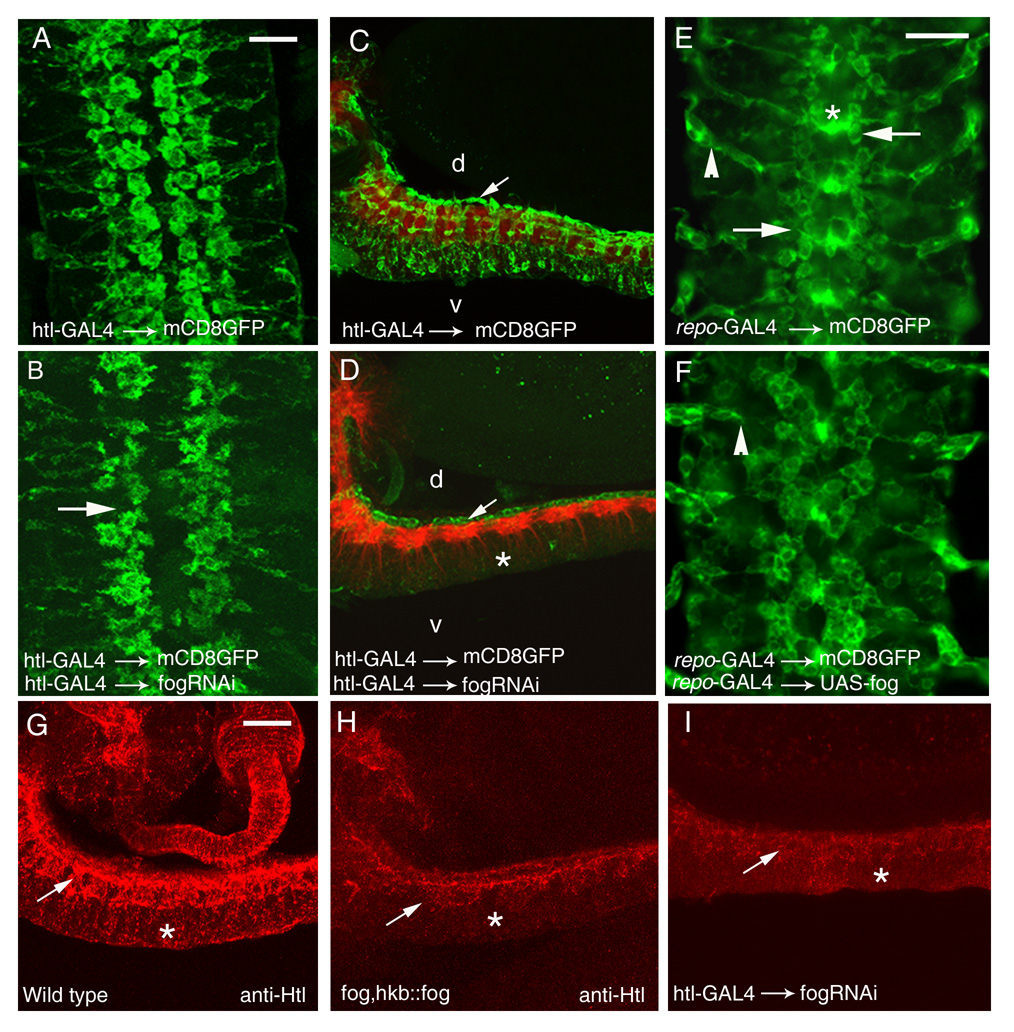
(A,B,E,F). Five segments of the CNS in late stage 16 embryos are shown, stained with anti-GFP to reveal the morphology of mCD8-GFP expressing glia; anterior is up. (C–D) and (G–I) are lateral views of the CNS; anterior is to the left and dorsal is up.
(A) Htl-GAL4::UAS-mCD8GFP. The longitudinal glia that express htl are organized in two rows on either side of the midline. The cells appear flat and spread out.
(B) Htl-GAL4::UAS-mCD8GFP, UAS-dsfogRNA (glial fog RNAi). The glial lattice is disrupted, with gaps appearing in glial rows (arrow). Glia appear smaller and rounder, and fingerlike projections are observed around their edges; the gap between rows at the midline is wider, primarily because the glia are smaller.
(C and D) Lateral view of the ventral nerve cord of late stage 16 embryos stained with mAb BP102 (red), which marks the axon scaffold, and anti-GFP (green) to reveal the glial extensions of mCD8-GFP expressing glia. d, dorsal; v, ventral.
(C) Htl-GAL4::UAS-mCD8GFP. Arrow indicates the longitudinal glial cell bodies overlying the axon scaffold (red). Glial processes are seen enveloping the axon bundles and glial staining is observed through the entire depth (d-v axis) of the nerve cord.
(D) Htl-GAL4::UAS-mCD8GFP, UAS-dsfogRNA (glial fog RNAi). Glial ensheathment of the nerve cord by longitudinal glia (arrow indicates cell bodies) is strongly affected. There is only a partial wrapping of glial processes around the red-labeled CNS axon scaffold. Glial staining in the region ventral to the axon layer is dramatically reduced (asterisk).
(E) Repo-GAL4::UAS-mCD8GFP. The longitudinal glia have a somewhat different appearance than when mCD8-GFP is driven with Htl-GAL4, but the two rows of longitudinal glia are clearly observed on either side of the midline (arrows). Glial processes spanning the midline (asterisks) are prominent, as are exit glia ensheathing the peripheral nerve roots (arrowhead). This driver provides better visualization of the exit glia than does Htl-GAL4.
(F) Repo-GAL4::UAS-mCD8GFP, UAS-fog (glial Fog overexpression). The longitudinal glial lattice is disorganized, as are the exit glia (arrowhead).
(G–I) Lateral view of the ventral nerve cord of late stage 16 embryos stained with anti-Htl antibody.
(G) Wild type. Htl staining is observed in a dorsal zone (arrow) made up of longitudinal glia that ensheath the neuropil, and a ventral zone (asterisk) made up of glia and glial processes that extend below the nerve cord.
(H) fog; hkb::fog. Htl staining in the dorsal zone (arrow) appears disorganized and its expression is almost absent in the ventral zone (asterisk). Overall Htl staining intensity is also reduced relative to wild-type.
(I) Htl-GAL4::UAS-mCD8GFP, UAS-dsfogRNA (glial fog RNAi). As in the fog mutant, Htl staining in the dorsal zone appears disorganized (arrow), Htl is very weak in the ventral zone (asterisk), and overall staining intensity is reduced.
Scale bar in A and E is 10 µm. Scale bar in A applies to panels A–D and G–I. Scale bar in E applies to panels E and F.
Fog staining in the CNS was carried out on live dissected and fixed embryos. The dissected embryos were fixed for 20 min. with 4% paraformaldehyde on ice. Blocking was done with PBTX containing 5% normal goat serum. Antibody staining was carried out using 1:100 dilution of the antibody. To detect secreted Fog protein, live dissected embryos were incubated with antibody for 2 hrs. at room temperature. The antibody was diluted in PBS containing 0.5% normal goat serum. After incubation with the antibody, the embryos were washed at least 3 times with PBS and fixed with 4% paraformaldehye. Subsequent detection of the antibody was carried out using standard procedures (Patel, 1994) embryos. The embryos were fixed for Alexa 488 or Alexa 568 secondary antibodies (Molecular Probes) were used for all fluorescent immunohistochemistry.
Images were processed using Adobe Photoshop 7. Quantitation of glial cell surface areas was carried out using ImageJ software (NIH).
Results
fog is expressed in longitudinal glia and neurons
To evaluate the expression pattern of the fog gene in the embryonic CNS, we performed in situ hybridization experiments using fog cDNA probes. We found that fog mRNA is uniformly expressed in germ band extended embryos (stages 10 and 11), but then becomes restricted to small groups of cells later in development.
fog mRNA expression within the CNS is most easily observed at stage 15 or later. Most of the cells expressing fog at high levels are located within narrow zones that flank the midline and extend from the anterior to the posterior of the embryo (Fig. 1A). These zones are two or three cells wide. Interestingly, however, not all cells within the zones express fog mRNA at levels above the detection threshold, and the pattern is not identical between segments or between embryos. This suggests that expression levels in individual cells might be determined in a stochastic manner.
To determine the identities of the fog-expressing cells, we double-stained embryos with fog probe (blue) and antibodies against glial and neuronal markers. We found that most of the prominent (high-level) fog-expressing cells in the CNS also stained for Repo, a transcription factor expressed in longitudinal glia (brown in Fig. 1B, arrows) (Xiong et al., 1994, Halter et al., 1995). There were also Repo-negative, fog-mRNA-positive cells within the CNS (Fig. 1B, asterisk). Some of these are located near the midline and might be midline glia, which stain with antibodies against Fog protein (Fig. 1C). Our results on expression of Fog protein (Fig. 1C–G, Supp. Fig. 1) indicate that it is expressed by some CNS and peripheral neurons as well; however, the levels of fog mRNA in neurons may be below the detection threshold for in situ hybridization. In the periphery, strong fog mRNA expression was observed in the scolopale and cap cells, which ensheath the dendrites of the sensory neurons in the chordotonal organs (Fig. 1E).
To study localization of Fog protein, we stained live-dissected stage 16 embryos using an antibody generated against full-length Fog (a gift from N. Fuse), and detected expression by immunofluorescence. We observed that Fog protein was localized to the scolopale and cap cells, consistent with the in situ hybridization results, and was also seen on the dendrites of chordotonal neurons (Fig. 1F). Background staining levels are high with this antibody. This is commonly observed with antibodies against secreted proteins, particularly when they must be used at high concentrations (1:100 in this case).
Within the CNS, the strongest staining with anti-Fog antibody was observed on the longitudinal axonal tracts, which are visualized with the axonal marker BP102 (Fig. 1C and C′). In order to confirm that this axonal staining represents genuine Fog protein, we utilized RNAi techniques, expressing fog double-stranded RNA (dsRNA) in embryos using a heat shock promoter-GAL4 driver (fog RNAi embryos; see below). Embryos that were heat shocked displayed decreased axonal staining with anti-Fog (Fig. 1D and 1D’), while those that were not subjected to heat shock showed strong Fog expression in the CNS.
We also expressed fog dsRNA with pan-neuronal (Elav) and longitudinal glial (Repo) drivers, and found that in neuronal fog RNAi embryos, CNS axonal staining was reduced, while glial fog RNAi did not strongly affect staining. In the periphery, neuronal fog RNAi eliminated staining of the dendritic shafts of the chordotonal neurons (Supp. Fig. 1). Expression in the scolopale and cap cells, however, appears to be resistant to both neuronal and heat-shock driven RNAi. It is also resistant to glial RNAi (not shown); this is expected since the scolopale and cap cells do not express Repo.
In order to evaluate phenotypic effects observed with glial fog RNAi, we needed to demonstrate that glial RNAi could inhibit Fog protein expression. Since Fog staining in the CNS apparently derives from both glia and the neurons they ensheath, and Fog is a secreted protein, effects of glial RNAi on staining with anti-Fog antibody are likely to be obscured by neuronal Fog expression. We thus examined Fog protein staining in the retinal basal glia of the larval eye disc and optic stalk, where we had observed that glial Fog expression can be spatially distinguished from expression in nearby neurons. We compared Fog staining between control larvae and glial fog RNAi larvae, and observed that staining in retinal basal glia (visualized with Repo-GAL4-driven mCD8-GFP) was reduced or eliminated in RNAi larvae (Supp. Fig. 1). The residual punctate staining in the glial region in the RNAi larvae probably represents the usual background observed with anti-Fog antibody.
Since Repo-positive glia comprise most of the high-level fog mRNA-expressing cells in the CNS, we wondered whether some of this axonal Fog might be transferred from glia to the axons. To examine this question, we stained gcm7–4 mutant embryos (Jones et al., 1995), in which most CNS glia are absent, with anti-Fog. We found that axonal Fog staining was unchanged (data not shown). Together with the RNAi data, these results indicate that axonal Fog is at least partially derived from Fog expressed in neurons.
The N terminus of the predicted 730 amino acid (aa) Fog protein resembles a signal sequence, and it has no obvious transmembrane domain. This is consistent with the prediction that Fog is a secreted ligand (Costa et al., 1994). It has also been demonstrated that Fog is transported through the secretory pathway (Dawes-Hoang et al., 2005). However, it has never been directly shown that Fog is extracellularly localized. To determine if Fog protein can be detected at or outside of the cell surface, we carried out immunofluorescent staining of unfixed live-dissected embryos in the absence of detergent. We observed expression of Fog on scolopale cells (Fig. 1F), indicating that Fog is indeed secreted or localized to the cell surface, at least in the periphery. Within the CNS, however, the high background staining observed under these conditions prevented us from clearly determining whether axonal Fog is also extracellular.
Reduction or overexpression of Fog in glia affects morphology and axon ensheathment
The results described above (Fig. 1) show that within the embryonic CNS fog mRNA is expressed primarily by Repo-positive glia. To evaluate the function of Fog in these glia, and to separate glial fog LOF phenotypes from those conferred by loss of Fog in neurons (see below), we used glial drivers to coexpress fog dsRNA together with mCD8-GFP, a membrane-tethered form of GFP, so that we could directly visualize glial morphology. Several independent UAS-fog dsRNA transgenic lines were examined, and all produced the same phenotypes.
To confirm the results obtained using RNAi techniques, we also examined fog LOF mutants in which early embryonic lethality caused by lack of PMG invagination (the folded phenotype) is rescued via expression of Fog in the posterior midgut region (PMG) from the huckebein (hkb) promoter (fog, hkb::fog; a gift of Maria Leptin). These embryos have near normal morphology at later stages of development (M. Leptin, personal communication). In particular, the ventrally located CNS develops normally in these embryos.
When Fog dsRNA was expressed using heartless (Htl)-GAL4 or Repo-GAL4 drivers (glial fog RNAi), we observed distinct changes in the positioning and shape of the longitudinal glia. In wild-type embryos, longitudinal glia expressing both Htl and Repo are organized in two rows on either side of the CNS midline (Fig. 2A) (Halter et al., 1995, Shishido et al., 1997). In glial fog RNAi embryos, gaps were observed in these rows (Fig. 2B, arrow). The glial cells appeared smaller and rounder than normal, and their outlines were irregular, often bearing tiny finger-like projections. The distance between the inner glial rows was also greater than in wild-type. To quantitate these phenotypes, we measured the sizes of the GFP-positive glial profiles, and observed that glial surface area was reduced by about 30% relative to controls (from 131 µm² (n=51) to 94 µm² (n=110)).
To see if the change in glial shape also affected glial wrapping of axons, we examined the CNS of glial fog RNAi and fog, hkb::fog embryos in side view. The glial fog RNAi embryos were co-stained with anti-GFP (in green) and the axon marker BP102 (in red). A series of confocal images (z stacks) were taken for both wild-type and RNAi embryos. Upon superimposing the images, we found that in glial fog RNAi embryos, the glial processes fail to extend ventrally and do not ensheath the axons (Figs. 2C–D). Glial staining is localized to the dorsal edge of the CNS, while in control (Htl-GAL4 × UAS-mCD8-GFP) embryos it extends ventrally beyond the axon layer.
Glial processes were also visualized with anti-Htl in both glial fog RNAi and fog, hkb::fog embryos. The normal Htl expression pattern is similar to that of Htl-GAL4-driven mCD8GFP (Fig. 2G). When fog expression is reduced, Htl-positive glial processes were not observed ventral to the CNS axon layer, and a disorganized pattern of axonal ensheathment was observed (Figs. 2H,I). Overall Htl staining intensity was also reduced. This is interesting, since the fog phenotype we observed resembles the previously described Htl LOF phenotype (Shishido et al., 1997).
To carry out overexpression experiments, we generated UAS-fog transgenes bearing the entire fog cDNA cloned into the pUAST vector. We confirmed that Fog protein was overexpressed in these embryos. Several independent transgenic lines were examined, and all had the same phenotypes. While glial fog RNAi animals survive to adulthood, animals in which Fog is overexpressed using glial drivers die as 1st or 2nd instar larvae. Examination of glial morphology in Repo-GAL4::UAS-mCD8GFP, UAS-fog embryos showed that they had severe glial defects. The longitudinal glia had a disorganized and clumped appearance, and extension of glial processes appeared abnormal (Figs. 2E–F).
Loss of neuronal Fog causes axon guidance defects
Our examination of Fog protein expression and its inhibition by RNAi indicates that Fog is expressed by at least some neurons (Fig. 1, Supp. Fig. 1). To evaluate whether neuronal Fog regulates axon guidance, we examined axons in two genetic contexts. First, we reduced Fog expression in neurons by driving fog dsRNA with the Elav-GAL4 driver, which is expressed in all postmitotic neurons (neuronal fog RNAi). Several independent UAS-fog dsRNA transgenic lines were examined, and all produced the same phenotypes. Second, we examined fog, hkb::fog embryos.
In the abdominal segments of wild-type embryos, motor axons exit the CNS via the SN (segmental nerve) and ISN (intersegmental nerve) roots. These then split into 5 different nerves, which innervate 30 body wall muscle fibers (Keshishian et al., 1996). These can all be visualized by staining with the 1D4 mAb (Van Vactor et al., 1993). SNa and SNc emerge from the SN root, while the ISN, ISNb and ISNd arise from the ISN root. The SNa nerve shows a characteristic pattern of bifurcation. The bifurcation occurs at the dorsal edge of muscle 12, at a point between muscles 22 and 23 (Figs. 3A–B). The anterior (or dorsal) SNa branch innervates muscles 21–24, while the posterior (or lateral) branch innervates muscles 5 and 8.
Figure 3. Motor axon phenotypes in fog and Ptp52F mutants.
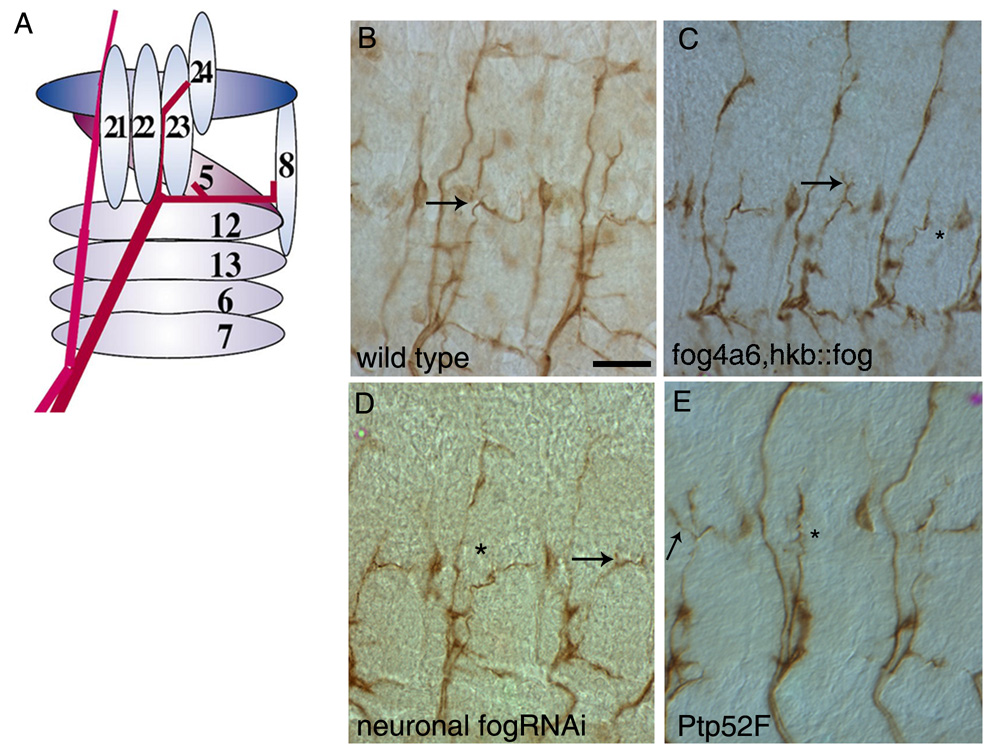
Late stage 16 embryo fillets stained with mAb1D4 (brown) to reveal motor axons and their projections. Two or three abdominal segments are shown in panels (B–E).
(A) Schematic representation of SNa bifurcation. The SNa nerve (red) branches at the dorsal edge of muscle 12. The anterior (dorsal) branch innervates muscles 21–24, and the posterior (lateral) branch innervates muscles 5 and 8. The pink nerve on the left is the ISN.
(B) Wild-type (Oregon R) embryo. Arrow indicates bifurcation point.
(C) fog4a6,hkb::fog mutant embryo. The posterior branch is missing in the right-hand segment (asterisk). An extra branch emerges from the anterior branch in the middle segment (arrow).
(D) Elav-GAL4::UAS-dsfogRNA (Neuronal fog RNAi) embryo. The anterior branch is missing in the middle segment (asterisk). In the right-hand segment, the anterior branch is truncated (arrow).
(E) Ptp52F18.3 mutant embryo. The posterior branch is missing in the middle segment (asterisk). An extra branch is seen in the left-hand segment (arrow). Scale bar in (B), 10 µm (applies to all panels).
Both fog4a6, hkb::fog and neuronal fog RNAi embryos had SNa motor axon guidance errors (scored in segments A2–A7). In most segments that displayed phenotypes, either the posterior or the anterior SNa branch was missing (Figs. 3C and D, asterisks). Occasionally, extra anterior branches were observed (Fig. 3C, arrow). In rare cases, the entire SNa appeared to stall at the bifurcation point. The penetrance of these defects was 13.2% and 12.5% in fog4a6,hkb::fog and neuronal fog RNAi embryos, respectively. We did not observe any motor axon guidance phenotypes in glial fog RNAi embryos.
Interestingly, the motor axon guidance defects in fog embryos were identical to those seen in mutants lacking expression of PTP52F (Fig. 3E) (Schindelholz et al., 2001) In Ptp52F embryos, the most common phenotype is also loss of the anterior or posterior SNa branch, and ectopic branches and stalling are observed at lower frequencies (Fig. 3E). The penetrance of these phenotypes is higher (~40% in null mutants) than in fog embryos. However, we do not know if our fog phenotypes represent the null condition. RNAi usually does not completely eliminate protein expression. Furthermore, fog4a6, hkb::fog may not be fog-null within the CNS, since hkb is expressed in a subset of CNS neuroblasts in each segment, and secreted Fog expressed by these cells could diffuse within the CNS (Chu-LaGraff etal., 1995, Bossing et al., 1996).
We also examined the three pairs of 1D4-positive longitudinal axon bundles in the CNS (Van Vactor et al., 1993) (Fig. 4A). In fog4a6,hkb::fog embryos, the outer 1D4-positive bundle was discontinuous, and occasional breaks and fusions were observed in the inner two bundles (Fig. 4B, arrow). Similar, but milder, phenotypes were observed in neuronal fog RNAi embryos (Fig. 4C). Glial fog RNAi embryos did not have CNS axonal phenotypes.
Figure 4. CNS phenotypes in fog and Ptp52F mutants.
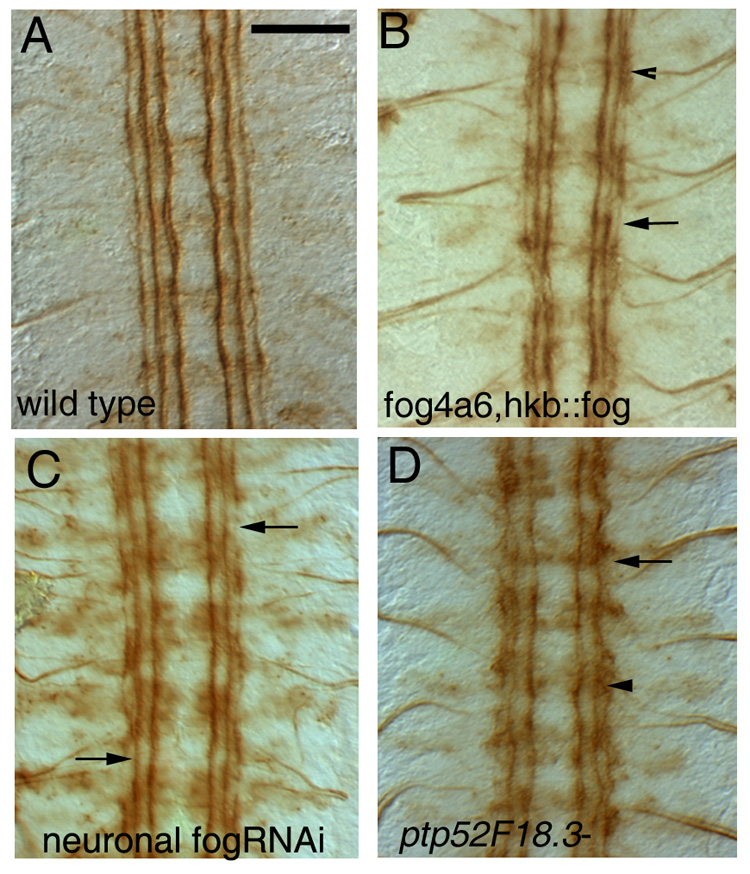
Each panel shows four segments of a late stage 16 embryo fillet stained with mAb 1D4 (brown); anterior is up. Scale bar in (A), 10um (applies to all panels).
(A) Wild-type (Oregon R) embryo. Note the three straight 1D4-positive longitudinal axon bundles on each side of the midline.
(B) fog4a6,hkb::fog mutant embryo. There are breaks in the outer bundle (arrows), and fusion of bundles are sometimes observed (arrowhead).
(C) Elav-GAL4::UAS-dsfogRNA (neuronal fog RNAi) embryo. A similar, but usually milder phenotype, is observed, in which the outer bundle is discontinuous (arrows).
(D) Ptp52F18.3 mutant embryo. The outer and middle 1D4-positive bundles are both disorganized. The outer bundle is also discontinuous (arrow) and is often fused to the middle bundle (arrowhead).
Interestingly, the fog4a6,hkb::fog CNS axonal phenotype also resembles that seen in Ptp52F mutants (Schindelholz et al., 2001). However, in Ptp52F mutants there is more disorganization of the outer 1D4 bundle, and it is often fused with the middle bundle (Fig 4D, arrowhead).
PTP52F is epistatic to Fog
Overexpression of Fog in neurons using Elav-GAL4 produced strong CNS defects. The three pairs of 1D4-positive longitudinal axon bundles in the CNS displayed multiple breaks and fusions, and failed to extend along straight pathways (Fig. 5B, arrowhead). Ectopic midline crossing by 1D4-positive longitudinal axons was observed in 27% of segments (Fig. 5B, arrow). In embryos carrying two copies of the GAL4 driver, the percentage of segments with ectopic midline crossing increased to 44% (Fig. 5F). SNa defects were also observed (Table 1).
Table 1.
SNa motor axon phenotypes in fog mutants
| GENOTYPE | Number of hemisegments | Total % SNa defects |
|---|---|---|
| elav-GAL4 × UAS-fog dsRNA (RT) | 278 | 12.5% |
| fog4a6,hkb::fog, (RT) | 356 | 13.2% |
| elav-GAL4 × UAS-fog, (25 deg). | 382 | 16.75% |
| elavc155-GAL4; elav-GAL4 × UAS-fog, (25 deg). | 398 | 20.6% |
| ctaRC10/ctaRC10, (RT). | 215 | 44.65% |
Abdominal segments A2–A7 were scored for SNa guidance defect.
Glial Fog overexpression also caused axon guidance defects, producing ectopic midline crossing phenotypes like those seen for neuronal Fog overexpression embryos, and with a similar penetrance (23%, Figs. 5E–F). These data suggest that excess secreted Fog within the CNS can affect axons in a similar manner regardless of whether it is expressed by neurons or by glia.
To see if PTP52F overexpression causes effects like those seen for Fog overexpression, we used Elav-GAL4 to drive expression from a UAS-bearing P element [EP element; Rorth et al., 1998] inserted immediately upstream of the Ptp52F gene (a gift of Florenci Serras). This produced CNS axon guidance defects that resembled those caused by excess Fog (Fig. 5C). The 1D4-positive axon bundles had breaks and defasiculated regions, and ectopic midline crossing by longitudinal axons was also observed in 18% (n=161) of segments (Fig. 5F).
These results show that fog and Ptp52F have similar LOF phenotypes in in the neuromuscular system and CNS. They also have similar CNS overexpression phenotypes. Because of these phenotypic resemblances, we conducted an epistasis analysis to see if Fog and PTP52F might function within the same signaling pathway. We found that removal of Ptp52F function from Fog overexpression embryos resulted in a strong suppression of the CNS phenotype (Figs. 5D,F). Only 3% of segments had ectopic midline crossing, as compared to 44% for Fog overexpression embryos with a wild-type Ptp52F background (embryos with two copies of neuronal GAL4 driver). For embryos with one copy of the driver, the phenotype was suppressed from 27% to 5%. The longitudinal tracts also looked more normal in Fog overexpression, Ptp52F embryos, resembling those seen in Ptp52F single mutants. These results indicate that the generation of ectopic midline crossing phenotypes by excess neuronal Fog signaling requires PTP52F, and suggests that this RPTP functions downstream of Fog in CNS neurons.
Discussion
In this paper, we show that the secreted cell signal Fog, which is necessary for VF formation in early embryos, also functions during development of the nervous system. Fog is expressed by both glia and neurons within the CNS (Fig. 1, Supp. Fig. 1.), and phenotypic analysis indicates that it has distinct functions in the two cell types. The level of glial Fog expression is a critical element in regulating migration of glial cell bodies, extension of processes, and ensheathment of axons (Fig. 2). Autocrine Fog reception by VF cells regulates cytoskeletal morphology and thereby cell shape (Dawes-Hoang et al., 2005), and Fog might function in a similar manner to control glial morphogenesis.
Reduction of Fog in neurons produces subtle axon guidance phenotypes affecting both motor neurons and CNS interneurons (Fig. 3,Fig. 4). Overexpression of Fog in neurons produces strong CNS phenotypes in which longitudinal axons abnormally cross the midline (Fig. 5). The same phenotypes can be produced by overexpressing Fog in CNS longitudinal glia, which are in apposition to the axons. This results suggests that glial Fog causes cytoskeletal changes that alter axon guidance in neurons, implicating Fog as an exocrine as well as an autocrine signal during nervous system development.
Studies of Fog signaling during gastrulation have indicated that the cytoskeletal changes produced by autocrine Fog involve maternal Cta and RhoGEF2, and nonmuscle myosin II (Dawes-Hoang et al., 2005). We tested whether these components participate in Fog signaling in the nervous system by examining the zygotic phenotypes of cta and RhoGEF2 mutants (germline clones do not develop to this stage). Cta may also be involved in Fog signaling during nervous system development, because we found that cta zygotic mutant embryos display the same defects in the CNS and neuromuscular system as do fog embryos (Table 1). RhoGEF2 mutants, however, have no visible nervous system defects (data not shown). Myosin II (zipper) mutant embryos have a variety of generalized defects that preclude analysis of specific axon guidance phenotypes.
Fog is an autocrine signal required for glial morphogenesis
Most of the cells in the CNS of late embryos that express fog mRNA at high levels are Repopositive longitudinal glia (Fig. 1). These glia are required for normal morphogenesis of the CNS axon tracts (Hidalgo et al., 1995, Hidalgo et al., 2000); but we did not observe CNS axon phenotypes when Fog expression was reduced in glia. To evaluate Fog's functions in the glia, we examined glial morphology directly using a membrane-associated GFP marker. When Fog expression is reduced in glia, glial processes fail to extend normally and ensheath CNS axons (Fig. 2). There are gaps in the regular array of glia, glial surface areas are smaller than in wild-type, and the glia have a rounded appearance. These changes in cell shape could involve nonmuscle myosin.
Overexpression of Fog in glia confers lethality during early larval phases. Glial morphogenesis is affected by overexpression, but the phenotypes are different from those seen when Fog is reduced. Glia appear to have normal shapes, but the glial lattice is quite disorganized. In wild-type embryos, lines of glia define the positions of the longitudinal tracts, commissural tracts, and peripheral nerves; these regular arrays are not observed in Fog glial overexpression embryos (Fig. 2). Thus, both reduction and elevation of glial Fog produces a disorganized glial lattice, suggesting that a precise level of the Fog signal is necessary for normal glial development.
Potential relationships between Fog and PTP52F
The Fog receptor has not been identified, although it is speculated to be a GPCR because of the requirement of the G protein alpha subunit Cta for Fog signaling (Morize et al., 1998). However, existing genetic data do not show that Fog directly activates a GPCR; they are also consistent with models in which Fog regulates signaling through a GPCR-Cta pathway in an indirect manner by interacting with a non-GPCR receptor.
PTP52F, like most RPTPs, is an 'orphan receptor'. The motivation to conduct the experiments described in this paper arose from our observations that fog and Ptp52F embryos display similar VF phenotypes (Supp. Fig. 2), and that PTP52F is expressed in ventral furrow cells during the gastrulation phase (Schindelholz et al., 2001). Based on these results, we wondered whether PTP52F could be the elusive Fog receptor.
PTP52F is required for axon guidance in the embryonic CNS and neuromuscular system. Thus, to examine whether Fog and PTP52F might be part of the same signaling pathway, we examined fog axon guidance phenotypes and studied genetic interactions between the two molecules. Our data show that fog and Ptp52F have similar LOF and GOF phenotypes in the CNS (Fig. 4,Fig. 5). In the neuromuscular system, LOF mutations in both genes cause SNa bifurcation phenotypes (Fig. 3). Our definition of a fog GOF CNS phenotype allowed us to perform an epistasis experiment, and this showed that PTP52F is required for signaling downstream of Fog, at least in the context of this phenotype (Fig. 5).
The genetic results we obtained indicate that PTP52F is involved in reception of the Fog signal by neurons, but do not prove that PTP52F is a Fog receptor. The results could also be explained if PTP52F positively regulates signaling through a Fog-GPCR-Cta pathway. For example, GPCRs are phosphorylated (on serine or threonine residues) and internalized; the activities of the relevant kinases and/or the proteins involved in internalization could be modulated by tyrosine phosphorylation. Tyrosine phosphorylation could also regulate effectors downstream of the G protein Cta.
We have been unable to demonstrate a direct biochemical interaction between the PTP52F extracellular domain and several versions of the Fog protein. However, Fog might be processed in vivo to create a functional ligand, and this processing does not occur in heterologous systems. In our experiments, we found that Fog tagged at its N-terminus is secreted from insect cells when expressed using the baculovirus system, but the protein is degraded to produce a ladder of bands ranging in size from ~100 kD (the predicted size of glycosylated full-length Fog) to <20 kD. Fog tagged at its C terminus cannot be detected at all. Fog fused near its C terminus to human placental alkaline phosphatase (Fog-AP) can be expressed as a mixture of apparently full-length and degraded forms, but none of these proteins bound detectably to the tagged PTP52F extracellular domain (A.R., P.M. Snow, and K.Z., unpublished results). Taken together, our data suggest that the C terminal region of Fog is subject to degradation, and that full-length Fog is unstable. There are several dibasic sequences in Fog which could represent proteolytic cleavage sites, and Wieschaus and colleagues (Costa et al., 1994) have previously proposed that Fog could be processed in vivo to generate an active fragment that binds to the receptor. In the CNS, such a fragment might derive from the middle region of Fog, because we observed that antisera against full-length Fog stain late embryos (Fig. 1, Supp. Fig. 1), while antisera against the first 300 amino acids of Fog (Dawes-Hoang et al., 2005) do not.
Supplementary Material
Panels A–C show 4 segments each of a stage 16 embryo kept at room temperature, dissected live and stained with anti-Fog antibody (from N. Fuse). Panels D–F show expression of Fog in lateral chordotonal organs. Anterior is up in A–C, and to the left in D–F. Panels G–L show retinal basal glia in 3rd instar larval eye discs stained with anti-Fog (from the Wieschaus lab; red) and anti-GFP (green).
(A) Fog staining in the ventral nerve cord of wildtype embryo. The highest levels of Fog expression are observed on the axon tracts (arrow) and the midline glia (asterisk).
(B) Elav-GAL4::UAS-fogdsRNA (neuronal fog RNAi). The level of Fog staining is reduced on the axon tracts, which are barely visible (arrow). Midline glial staining is still detectable (asterisk), but is reduced in intensity, suggesting that some of the Fog on the surfaces of the midline glia is secreted from nearby neurons.
(C) Repo-GAL4::UAS-fogdsRNA (glial fog RNAi). The CNS axon tracts (arrow) and midline glia (asterisk) are still visible (arrow), although staining may be somewhat weaker than in wild-type.
(D) Wild-type embryo. Fog expression is observed on the dendritic shafts of the sensory neurons (arrow), the scolopale (yellow arrowhead) and the cap cells (white arrowhead).
(E) Neuronal fog RNAi. Fog staining on the dendritic shaft is undetectable (arrow) but the staining in the scolopale (yellow arrowhead) and cap cells (white arrowhead) is unaltered.
(F) hs-GAL4::UAS-fogdsRNA (heat-shock fog RNAi). Fog staining of the dendritic shaft (arrow) is undetectable, as in (E). Fog expression in the scolopale (yellow arrowhead) and cap cells (white arrowhead) is apparently resistant to RNAi. Scale bar: 10um (A,B,C), 10um (D,E,F).
(G–I) Eye disc from 3rd instar RepoGAL4::UAS-mCD8GFP larva. (J–L) Eye disc from 3rd instar Repo-GAL4:: UAS-mCD8GFP, fog dsRNA larva (glial RNAi). Panels (G) and (J) show GFP staining in the retinal basal glia (RBG, arrows). Scale bar in (G) corresponds to 20µm and applies to all panels. Panel (H) shows Fog staining in the region of the RBG. We could detect Fog expression in the RBG with either the antibody against the full length protein from N.Fuse or the antibody from the Wieschaus lab against the N terminus. These panels show staining with the antibody against the N terminus. (I) A merged image of panels (G) and (H) showing that Fog is expressed by the RBG. (J) Eye disc from a glial RNAi larva. GFP expression marks the RBG. (K) Fog staining of the RBG is significantly reduced (arrow). Some residual punctate staining is observed which probably represents the background staining usually observed with this antibody. (L) A merged image of panels (J) and (K).
The ventral aspect of stage 6 whole-mount embryos stained with anti-Twist antibody using HRP immunohistochemistry for detection. (A,C,E) Whole embryos (anterior to the left); (B,D,F) High-magnification view of ventral furrow. (A,B) Wild-type. The furrow appears as a straight line in (B) (arrow). (C–D) Ptp52F18.3 mutant embryo. The Twist-expressing band is wavy and a straight furrow line is not visible in (D) (arrow marks approximate furrow position). (E–F) fog4a6 mutant embryo. The phenotype is similar to that of Ptp52F. Scale bars: 10 µm (A,C,E), 10 µm (B,D,F).
Acknowledgements
We are very grateful to the Wieschaus lab and Naoyuki Fuse for their generous gift of the Fog antibody and to Maria Leptin for sharing the fog4a6, hkb::fog flies prior to publication. We also thank Florenci Serras for EP52F; the Caltech Biological Imaging Centre and the Gonda Imaging Centre, UCLA for use of their confocal microscopes. Aloisia Schmid and Benno Schindelholz (former Zinn group members) discovered the Ptp52F VF phenotype. We thank them and the other present and former members of the Zinn group for helpful discussions. A.R. particularly thanks Dr. George Jackson, UCLA, in whose lab some part of this work was done. This work was supported by an NIH RO1 grant, NS28182, to K. Zinn, and by the Gosney fellowship from Caltech to A.R.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bossing T, Technau GM, Doe CQ. huckebein is required for glial development and axon pathfinding in the neuroblast 1-1 and neuroblast 2-2 lineages in the Drosophila central nervous system. Mech Dev. 1996;55:53–64. doi: 10.1016/0925-4773(95)00490-4. [DOI] [PubMed] [Google Scholar]
- Chu-LaGraff Q, Schmid A, Leidel J, Bronner G, Jackle H, Doe CQ. huckebein specifies aspects of CNS precursor identity required for motoneuron axon pathfinding. Neuron. 1995;15:1041–1051. doi: 10.1016/0896-6273(95)90093-4. [DOI] [PubMed] [Google Scholar]
- Costa M, Sweeton D, Wieschaus E. Gastrulation in Drosophila: cellular mechanisms of morphogenetic movements. In: Bate M, Martinez-Arias A, editors. The Development of Drosophila. New York: Cold Spring Harbor Laboratories Press; 1993. pp. 425–465. [Google Scholar]
- Costa M, Wilson ET, Wieschaus E. A putative cell signal encoded by the folded gastrulation gene coordinates cell shape changes during Drosophila gastrulation. Cell. 1994;76:1075–1089. doi: 10.1016/0092-8674(94)90384-0. [DOI] [PubMed] [Google Scholar]
- Dawes-Hoang RE, Parmar KM, Christiansen AE, Phelps CB, Brand AH, Weichaus EF. folded gastrulation, cell shape change and the control of myosin localization. Development. 2005;132:4165–4178. doi: 10.1242/dev.01938. [DOI] [PubMed] [Google Scholar]
- Hidalgo A, Urban J, Brand AH. Targeted ablation of glia disrupts axon tract formation in the Drosophila CNS. Development. 1995;121:3703–3712. doi: 10.1242/dev.121.11.3703. [DOI] [PubMed] [Google Scholar]
- Hidalgo A, Booth GE. Glia dictate pioneer axon trajectories in the Drosophila embryonic CNS. Development. 2000;127:393–402. doi: 10.1242/dev.127.2.393. [DOI] [PubMed] [Google Scholar]
- Halter DA, Urban J, Rickert C, Ner SS, Ito K, Travers AA, Technau GM. The homeobox gene repo is required for the differentiation and maintenance of glia function in the embryonic nervous system of Drosophila melanogaster. Development. 1995;121:317–332. doi: 10.1242/dev.121.2.317. [DOI] [PubMed] [Google Scholar]
- Jones BW, Fetter RD, Tear G, Goodman CS. glial cells missing: a genetic switch that controls glial versus neuronal fate. Cell. 1995;82:1013–1023. doi: 10.1016/0092-8674(95)90280-5. [DOI] [PubMed] [Google Scholar]
- Keshishian H, Broadie K, Chiba A, Bate M. The Drosophila neuromuscular junction: a model system for studying synaptic development and function. Annu Rev Neurosci. 1996;19:545–575. doi: 10.1146/annurev.ne.19.030196.002553. [DOI] [PubMed] [Google Scholar]
- Kraut R, Menon K, Zinn K. A gain-of-function screen for genes controlling motor axon guidance and synaptogenesis in Drosophila. Curr Biol. 2001;11:417–430. doi: 10.1016/s0960-9822(01)00124-5. [DOI] [PubMed] [Google Scholar]
- Lammel U, Saumweber H. X-linked loci of Drosophila melanogaster causing defects in the morphology of the embryonic salivary glands. Dev Genes Evol. 2000;210:525–535. doi: 10.1007/s004270000096. [DOI] [PubMed] [Google Scholar]
- Morize P, Christiansen AE, Costa M, Parks S, Wieschaus E. Hyperactivation of the folded gastrulation pathway induces specific cell shape changes. Development. 1998;125:589–597. doi: 10.1242/dev.125.4.589. [DOI] [PubMed] [Google Scholar]
- Parks S, Wieschaus E. The Drosophila gastrulation gene concertina encodes a G alpha-like protein. Cell. 1991;64:447–458. doi: 10.1016/0092-8674(91)90652-f. [DOI] [PubMed] [Google Scholar]
- Patel NH. Imaging neuronal subsets and other cell types in whole-mount Drosophila embryos and larvae using antibody probes. In: Fryberg E, Goldstein LSB, editors. Drosophila melaogaster: Practical uses in Cell and Molecular Biology. Vol 44. San Diego, CA: Academic Press; 1994. pp. 446–488. [DOI] [PubMed] [Google Scholar]
- Rorth P, Szabo K, Bailey A, Laverty T, Rehm J, Rubin GM, Weigmann K, Milan M, Benes V, Ansorge W, Cohen SM. Systematic gain-of-function genetics in Drosophila. Development. 1998;125:1049–1057. doi: 10.1242/dev.125.6.1049. [DOI] [PubMed] [Google Scholar]
- Schindelholz B, Knirr M, Warrior R, Zinn K. Regulation of CNS and motor axon guidance in Drosophila by the receptor tyrosine phosphatase DPTP52F. Development. 2001;128:4371–4382. doi: 10.1242/dev.128.21.4371. [DOI] [PubMed] [Google Scholar]
- Shishido E, Ono N, Kojima T, Saigo K. Requirements of DFR1/Heartless, a mesoderm-specific Drosophila FGF-receptor, for the formation of heart, visceral and somatic muscles, and ensheathing of longitudinal axon tracts in CNS. Development. 1997;124:2119–2128. doi: 10.1242/dev.124.11.2119. [DOI] [PubMed] [Google Scholar]
- Sweeton D, Parks S, Costa M, Wieschaus E. Gastrulation in Drosophila: the formation of the ventral furrow and posterior midgut invaginations. Development. 1991;112:775–789. doi: 10.1242/dev.112.3.775. [DOI] [PubMed] [Google Scholar]
- Van Vactor D, Sink H, Fambrough D, Tsoo R, Goodman CS. Genes that control neuromuscular specificity in Drosophila. Cell. 1993;73:1137–1153. doi: 10.1016/0092-8674(93)90643-5. [DOI] [PubMed] [Google Scholar]
- Xiong WC, Okano H, Patel NH, Blendy JA, Montell C. repo encodes a glial-specific homeo domain protein required in the Drosophila nervous system. Genes Dev. 1994;8:981–994. doi: 10.1101/gad.8.8.981. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Panels A–C show 4 segments each of a stage 16 embryo kept at room temperature, dissected live and stained with anti-Fog antibody (from N. Fuse). Panels D–F show expression of Fog in lateral chordotonal organs. Anterior is up in A–C, and to the left in D–F. Panels G–L show retinal basal glia in 3rd instar larval eye discs stained with anti-Fog (from the Wieschaus lab; red) and anti-GFP (green).
(A) Fog staining in the ventral nerve cord of wildtype embryo. The highest levels of Fog expression are observed on the axon tracts (arrow) and the midline glia (asterisk).
(B) Elav-GAL4::UAS-fogdsRNA (neuronal fog RNAi). The level of Fog staining is reduced on the axon tracts, which are barely visible (arrow). Midline glial staining is still detectable (asterisk), but is reduced in intensity, suggesting that some of the Fog on the surfaces of the midline glia is secreted from nearby neurons.
(C) Repo-GAL4::UAS-fogdsRNA (glial fog RNAi). The CNS axon tracts (arrow) and midline glia (asterisk) are still visible (arrow), although staining may be somewhat weaker than in wild-type.
(D) Wild-type embryo. Fog expression is observed on the dendritic shafts of the sensory neurons (arrow), the scolopale (yellow arrowhead) and the cap cells (white arrowhead).
(E) Neuronal fog RNAi. Fog staining on the dendritic shaft is undetectable (arrow) but the staining in the scolopale (yellow arrowhead) and cap cells (white arrowhead) is unaltered.
(F) hs-GAL4::UAS-fogdsRNA (heat-shock fog RNAi). Fog staining of the dendritic shaft (arrow) is undetectable, as in (E). Fog expression in the scolopale (yellow arrowhead) and cap cells (white arrowhead) is apparently resistant to RNAi. Scale bar: 10um (A,B,C), 10um (D,E,F).
(G–I) Eye disc from 3rd instar RepoGAL4::UAS-mCD8GFP larva. (J–L) Eye disc from 3rd instar Repo-GAL4:: UAS-mCD8GFP, fog dsRNA larva (glial RNAi). Panels (G) and (J) show GFP staining in the retinal basal glia (RBG, arrows). Scale bar in (G) corresponds to 20µm and applies to all panels. Panel (H) shows Fog staining in the region of the RBG. We could detect Fog expression in the RBG with either the antibody against the full length protein from N.Fuse or the antibody from the Wieschaus lab against the N terminus. These panels show staining with the antibody against the N terminus. (I) A merged image of panels (G) and (H) showing that Fog is expressed by the RBG. (J) Eye disc from a glial RNAi larva. GFP expression marks the RBG. (K) Fog staining of the RBG is significantly reduced (arrow). Some residual punctate staining is observed which probably represents the background staining usually observed with this antibody. (L) A merged image of panels (J) and (K).
The ventral aspect of stage 6 whole-mount embryos stained with anti-Twist antibody using HRP immunohistochemistry for detection. (A,C,E) Whole embryos (anterior to the left); (B,D,F) High-magnification view of ventral furrow. (A,B) Wild-type. The furrow appears as a straight line in (B) (arrow). (C–D) Ptp52F18.3 mutant embryo. The Twist-expressing band is wavy and a straight furrow line is not visible in (D) (arrow marks approximate furrow position). (E–F) fog4a6 mutant embryo. The phenotype is similar to that of Ptp52F. Scale bars: 10 µm (A,C,E), 10 µm (B,D,F).


