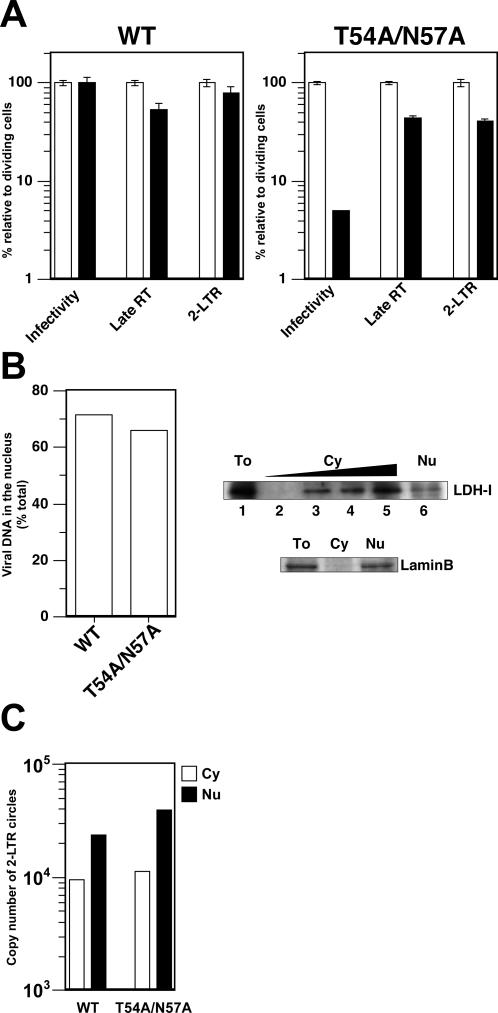
Figure 3. Reverse Transcription and Nuclear Import of Viral DNA in Nondividing Cells.
(A) Viral DNA synthesis and formation of 2-LTR circles. Dividing (shown in white bars) and nondividing HeLa cells (shown in black bars) prepared by aphidicolin treatment were infected with reporter virus constructs bearing the WT CA or T54A/N57A mutation. Virus infectivity was assessed by quantifying GFP-positive cells 2 d after infection. Relative infectivity is shown as percentage of infectivity in dividing cells. Total DNA was extracted 1 d after infection and used in real-time PCR to measure the copy number of late products of reverse transcription (RT) as well as that of 2-LTR circles. The data is shown as relative values where the amount in dividing cells for each different virus is set at 100%. Infections were done in triplicate and error bars indicate standard deviation. Two independent experiments were performed with similar results. One representative experiment is shown here.
(B) Nuclear localization of viral DNA. Nondividing cells infected with either WT HIV-1 or the CA mutant T54A/N57A were separated into cytoplasmic and nuclear fractions (left panel). DNA extracted from each fraction was used as template for real-time PCR to measure newly synthesized viral DNA. The efficiency of nuclear migration of viral DNA was examined by dividing the copy number of viral DNA in nuclear fractions by that in both fractions (cytoplasmic plus nuclear fractions). The data shown here is a representative of two independent experiments; although the ratio of viral DNA associated with nuclear fractions differed between two experiments, the amount of viral DNA associated with nuclear fraction remains the same between WT and the CA mutant. Controls with reverse transcriptase inhibitors showed that contamination by plasmid DNA accounted for less than 1% of the values (not shown). Western blotting analysis was performed to ensure the integrity of subcellular fractionation (right panels). Contamination was checked by checking a cytoplasmic protein, LDH-I (upper lanes) and a nuclar protein, lamin B (lower lanes). To, total cell lysates; Cy, cytoplasmic extract; Nu, nuclear lysates. Twenty migrograms of proteins were loaded on the gel except for the following dilutions. Cytoplasmic extract was diluted by 5-fold; 5-, 25-, and 125-fold dilutions correspond to lanes 4, 3, and 2 (upper lanes).
(C) Subcellular localization of 2-LTR circles. Copy numbers of 2-LTR circles in cytoplasmic (white boxes) and nuclear (black boxes) fractions were measured by real-time PCR. The same fractions as in the (B) were used in this PCR assay.