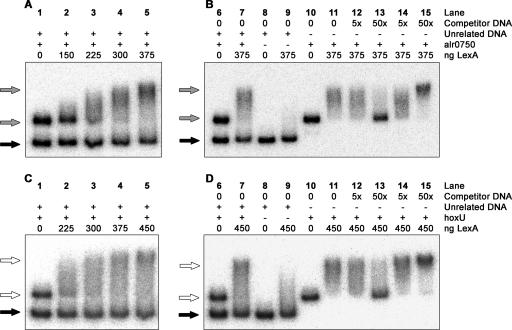
FIG. 5.
EMSAs with purified LexA from Nostoc sp. strain PCC 7120 and the two regulatory hox operon regions. The alr0750 (A and B) and hoxU (C and D) DNA fragments were generated by PCR using genomic DNA from Nostoc sp. strain PCC 7120, and the unrelated DNA fragment was generated by using pQE-30 vector (QIAGEN). The positions of the DNA fragments relative to the regulatory regions of the hox operons are shown in Fig. 2. EMSAs were first carried out with the specific alr0750 (A) and hoxU (C) fragments, incubated together with the unrelated DNA fragment without protein (lane 1) or together with increasing amounts of LexA (lanes 2 through 5). The unrelated DNA is indicated with a black arrow and the alr0750 and hoxU fragments and their retardations are indicated with gray and white arrows, respectively. EMSAs were carried out to demonstrate the specificities in the binding between Nostoc sp. strain PCC 7120 LexA and the alr0750 (B) and hoxU (D) fragments. The unrelated DNA was incubated with and without LexA, alone or together with the alr0750 or hoxU fragment (lanes 6 through 9). The alr0750 and hoxU fragments were also incubated together with a 5× or 50 × excess of either unlabeled specific (lanes 12 and 13) or nonspecific (lanes 14 and 15) DNA. The amount of LexA (in ng) used for each lane is indicated in the figure. A plus sign indicates that the fragment was included in the assay and a minus sign indicates that the fragment was excluded from the assay.