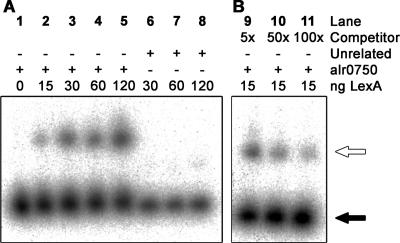
FIG. 6.
EMSAs with purified LexA from Synechocystis sp. strain PCC 6803 and the Nostoc sp. strain PCC 7120 regulatory hox operon region covered by the alr0750 DNA fragment. The fragments used were generated by PCR using genomic DNA from Nostoc sp. strain PCC 7120 (lanes 1 through 5 and 9 through 11) or pQE-30 vector from QIAGEN (unrelated DNA, lanes 6 through 8). The position of the DNA fragment relative to the regulatory region of the hox operon is shown in Fig. 2. The assays were carried out using 20 fmol of each labeled fragment incubated with 0, 15, 30, 60, or 120 ng purified LexA (lanes 1 through 5). Competition experiments were carried out with 15 ng LexA in the presence of 5×, 50 ×, and 100 × molar excesses of the unlabeled alr0750 fragment as competitor DNA (lanes 9 through 11). White arrows indicate unbound DNA fragments, and black arrows indicate a shift, i.e., LexA bound to the DNA fragment.