Abstract
siRNA delivery to cells offers a convenient and powerful means of gene silencing that bypasses several barriers met by gene delivery. However, nonviral vectors, and especially polymers, form looser complexes with siRNA than with plasmid DNA. As a consequence, exchange of siRNA for larger polymeric anions such as proteoglycans found outside cells and at their surface may occur and lower delivery. We show here that making siRNAs “gene-like,” via short complementary A5–8/T5–8 3′ overhangs, increases complex stability, and hence RNase protection and gene silencing in vitro up to 10-fold. After decomplexation in the cytoplasm, sticky siRNA (ssiRNA) concatemers fall apart. ssiRNAs are therefore not inducing antiviral responses, as shown by the absence of IFN-β production. Finally, transfection experiments in the mouse lung show that ssiRNA should be particularly suited to silencing with linear polyethylenimine in vivo.
Keywords: nonviral delivery, polyethylenimine, RNA interference
Drug development based on RNAi parallels antisense and antigene strategies, yet with a better prognosis (1, 2). Indeed, siRNAs outclass antisense oligonucleotides for gene expression knockdown in cell cultures. Moreover, RNAi occurs via a fast and efficient endogeneous machinery, whereas antigene approaches suffer from slow DNA triple-helix formation or strand invasion. Unfortunately, RNAi therapy shares with gene therapy the lack of efficient in vivo delivery techniques. But here again a better future is predicted, provided the drugs are small RNAs. Indeed, siRNAs and their complexes are expected to have much larger extracellular and intracellular diffusion coefficients than genes and RNAi is not hampered by the nuclear membrane.
Nonviral gene delivery reagents can be used also for siRNA delivery. Among in vivo delivery reagents, linear polyethylenimine (PEI) is being widely used for its versatility (3–5) and favorable efficiency/toxicity balance (6) due to efficient cargo translocation to the cytoplasm by the proton sponge effect (7, 8). However, complex formation and interaction of the cationic polymer with a gene or an siRNA differ by nature and strength. Indeed, a rigid two-turn double-helical siRNA cannot be “condensed,” complex formation lacks cooperativity, and electrostatic binding forces with PEI are reduced 100-fold. The major consequence of these differences may be unwanted exchange with large polyanions found outside cells. We reasoned that making siRNA “look like a gene,” e.g., by concatemerization, would restore the original delivery power of PEI. However, long dsRNAs trigger nonspecific gene shutdown. Concatemerization should therefore remain reversible and restricted to within the complex with PEI. We show here that siRNAs with short sticky overhangs (ssiRNAs) fulfill these requirements.
Results and Discussion
Cellular Fate of a Fluorescent siRNA When Delivered by a Cationic Lipid or a Cationic Polymer.
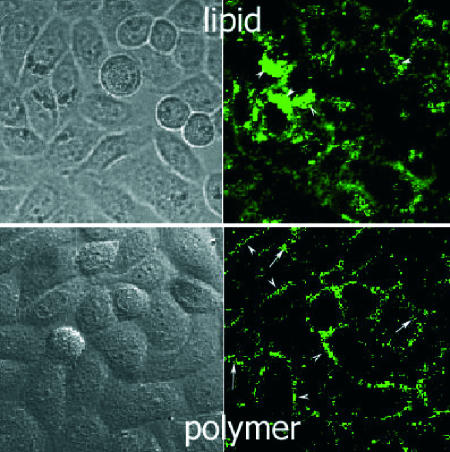
As a test of our working hypotheses, we followed delivery of a 6-FAM-labeled fluorescent siRNA to HeLa cells. Complexes were formed with a cationic lipid, jetSi-ENDO, and a cationic polymer, jetPEI. HeLa cells were incubated for 24 h with 100 nM complexed siRNA and observed by confocal microscopy.
With the cationic lipid, a section through the cells (Fig. 1) showed a distribution of discrete complexes similar to that observed for delivery of plasmid DNA, i.e., scattered intracytoplasmic spots with accumulation in the perinuclear region (Fig. 1, arrowheads). Most of these complexes are thought to be sequestered in endosomes/lysosomes as previously shown for plasmid DNA complexed to cationic lipids (9). The cationic polymer displayed a quite different pattern, with an essentially punctuate pericellular lining (Fig. 1, arrowheads) and some intracellular patches (Fig. 1, arrows). Free, diffuse, cytoplasmic siRNA was not observed, possibly as a consequence of fading upon decomplexation and dilution throughout the cytoplasm.
Fig. 1.
Confocal microscopy sections of HeLa cells transfected with a fluorescent siRNA complexed to a cationic lipid or a cationic polymer. Lipid complexes are found in the perinuclear lysosomal compartment (Upper, arrowheads). Polymer complexes are mostly found at the cell surface (Lower, arrowheads) and some times inside the cell (arrows).
The observed pericellular fluorescence can be explained by assuming that much of the siRNA is expelled from PEI/siRNA complexes by competition with polyanionic syndecans upon binding to the cell surface. Indeed, syndecan adhesion molecules are abundant on adherent cells where they act as receptors for cationic nonviral vectors (10). Binding of a polycationic particle leads to syndecan clustering at the cell surface, which in turn triggers actin-mediated endocytosis of the particle. According to the molecular weight (11) and degree of sulfatation of their heparan sulfate (HS) and chondroitine sulfate branches (12, 13), a rough estimate of the number of anionic charges per syndecan is of several hundreds. Competitive exchange between proteoglycans and the less-charged siRNA molecules could therefore occur in favor of the former remaining complexed to PEI.
Net siRNA cell entry is the resultant of endocytosis rate minus exchange rate before endosome closure. Exchange between nucleic acid and proteoglycan ionic polymers is probably faster within homogeneous complexes formed with another ionic polymer than within multilamellar lipid complexes (14) where lateral 2D diffusion is much less effective (15).
Challenging ssiRNA/PEI Complexes with Anionic Glycosaminoglycans or RNase.
Complex formation and release of siRNA from the complexes was studied with a model system using a 21-mer dsDNA by following ethidium bromide (EtdBr) fluorescence enhancement (16). Indeed, EtdBr intercalates between B-DNA base pairs, and radiative decay becomes favored over dipolar exchange with water. When DNA is complexed with polycations, EtdBr intercalation does not occur.
In typical transfection ratio conditions [5 μM DNA base pairs, jetPEI amine over DNA phosphate charge ratio (N/P) = 5], we found that addition of a 4-fold charge excess of chondroitin sulfate vs. dsDNA led to fast fluorescence increase. After 30 min, fluorescence had reached a plateau that was corresponding to release of 70% of the dsDNA oligonucleotide from the complexes. In agreement with the conclusions inferred from Fig. 1, no fluorescence increase was detected with the cationic lipid jetSi-ENDO complexes in standard transfection conditions (N/P = 6).
Direct visualization of siRNA by gel electrophoresis was also performed. Complex formation between siRNA and increasing amounts of PEI or cationic lipid was followed by agarose gel electrophoresis of the mixtures. As seen for plasmid DNA, the nucleic acid spot disappeared above N/P = 2 and complexes remained in the wells (data not shown). PEI/siRNA complexes (N/P = 5) formed with 500 nM siRNA were then incubated with increasing amounts of HS for 30 min. Electrophoresis showed siRNA to be released by HS in a concentration-dependent manner (Fig. 2Left). Total release was achieved above 20 HS/siRNA (wt/wt) equivalents of the polyanion. The siRNA used was a classical 19-mer dsRNA with 3′-(dT)2 overhangs. Extending the overhangs to (dT)8, i.e., increasing the overall siRNA anionic charge from 40 to 52, did not improve stability of the complexes in the presence of HS (Fig. 2 Right). Remarkably, complexes of a ssiRNA capable of forming noncovalent concatemers via (dA)8·(dT)8 bridges became resistant to exchange in these conditions (Fig. 2 Center). In similar HS competition conditions, plasmid DNA was also retained in the PEI/DNA complexes (data not shown), suggesting that the stability of ssiRNA complexes was caused by formation of concatemers. Concatemers were not detected by electrophoresis in the absence of PEI, because of low duplex overhang stability [the A8·T8 melting temperature is <10°C (17)] and possibly concatemers' size distribution. Indeed, large duplex stability increase in the presence of cationic polymers (18) or amphiphiles (19, 20) is well documented. It is therefore reasonable to assume that complex formation of many siRNA molecules with a polycationic PEI molecule or a liposome favors intracomplex encounter of siRNAs and shields their repulsive forces. The resulting gene-like concatemers have a much larger overall electrostatic interaction with the delivery vector, hence are less prone to exchange with other polyanions.
Fig. 2.
Gel electrophoresis of siRNA/PEI complexes incubated with increasing amounts of HS. Because of aggregation, siRNA/PEI complexes remain in the well (0 eq HS). Incubation for 30 min with 20–40 (wt/wt) eq HS releases siRNA (Left and Right), but not ssiRNA (Center), from the complexes.
PEI is efficiently protecting plasmid DNA, but much less so siRNA (21), from nuclease degradation. We therefore tested whether ssiRNA/PEI complexes would be less prone to endonuclease attack than siRNA/PEI complexes. Complexes were incubated with increasing amounts of RNase A for 30 min and loaded on an agarose gel with 0.65% SDS. The remaining dsRNA was quantitated by densitometry after EtdBr staining. siRNA degradation became visible with 5 μg of RNase and was complete with 100 μg of RNase. In contrast, ssiRNA remained intact within experimental error (±10%) in these conditions, suggesting that complex stability and chemical stability are linked.
Sticky Overhangs Enhance siRNA-Mediated Silencing with PEI.
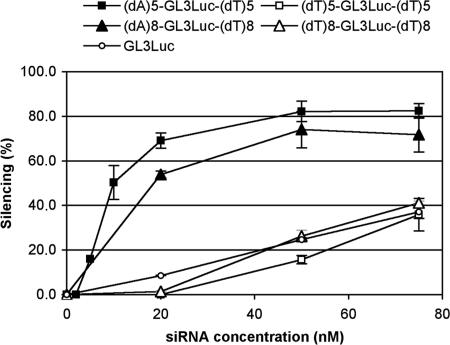
As a convenient cellular model to quantify endogeneous gene silencing, we chose the luciferase reporter protein that has a much shorter lifetime (3 h) (22) than GFP (23). To this end, human lung carcinoma A549 epithelial cells were stably transfected with pGL3Luc plasmid carrying the Photinus pyralis luciferase gene under control of simian virus 40 elements. A549Luc cells steadily expressed large amounts of luciferase [8 × 109 relative light units (RLU)/mg protein, 20 ng luciferase/mg protein]. The GL3Luc siRNA sense sequence 5′-CUUACGCUGAGUACUUCGA-(dT)2 has been described (24). Luciferase knockdown was assessed after 48-h incubation with various amounts of PEI/siRNA complexes. GL2Luc siRNA 5′-CGUACGCGGAAUACUUCGA-(dT)2 with three mismatches was used as negative control.
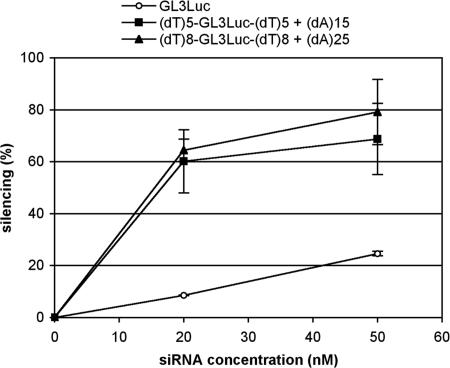
The effect of concatemer formation was assessed by comparing luciferase inhibition efficiencies of sticky (dA)5-GL3Luc-(dT)5 and (dA)8-GL3Luc-(dT)8 siRNAs with their cognate nonsticky (dT)5-GL3Luc-(dT)5 and (dT)8-GL3Luc-(dT)8 sequences at various concentrations. As shown in Fig. 3, the nonsticky siRNAs displayed silencing profiles similar to the classical GL3Luc with (dT)2 overhangs. In contrast, the two ssiRNAs showed much enhanced silencing at all concentrations. Silencing reached a 80% limit for 50 nM siRNA, presumably as a consequence of the large amounts of luciferase produced by these cells. Enhancement was estimated to ≈10-fold before approaching the saturation regime.
Fig. 3.
Endogeneous luciferase silencing with ssiRNA/PEI complexes requires 10-fold less RNA. A549Luc cells stably expressing GL3 luciferase were incubated with increasing concentrations of ssiRNAs complexed to PEI (N/P = 5). Control RNAs were the corresponding nonsticky siRNA of similar length and the classical GL3 siRNA with 3′(dT)2 overhangs. Luciferase inhibition was measured after 48 h.
To see whether similar effects could be observed with another gene and cell type, we targeted the intermediate filament protein vimentin mRNA in 3T3 fibroblasts. Silencing was assessed after 72 h vs. an irrelevant sequence by densitometric analysis of a Western blot. Protein levels were normalized to GAPDH. At 50 and 75 nM concentration, silencing with jetPEI/siRNA was ineffective (20 ± 20%). In contrast, ssiRNA led to robust silencing (70% and 90%) for these concentrations.
ssiRNA-Mediated Silencing Is Specific and Does Not Trigger Antiviral Responses.
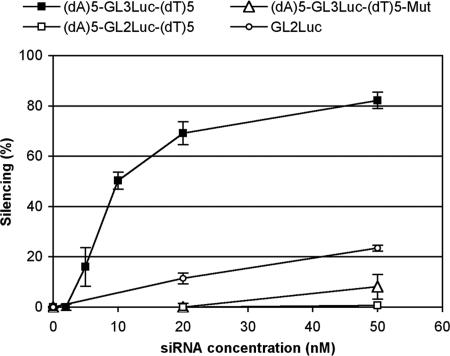
ssiRNA (dA)5-GL3Luc-(dT)5 silencing was compared with negative control (dA)5-GL2Luc-(dT)5 ssiRNA with three mismatches and to a single-mutation variant (dA)5-CUUACGCUAAGUACUUCGA-(dT)5 ssiRNA. As shown in Fig. 4, discrimination was as good as for classical siRNA. Moreover, control GL2 ssiRNAs did not elicit higher nonspecific silencing than GL2 siRNA, suggesting that ssiRNAs do not elicit the cellular responses described for long dsRNA.
Fig. 4.
ssiRNA-mediated silencing is sequence-specific. Sequences with three mismatches (○ and □) or with a single mismatch (■) in their GL3Luc sequence do not decrease endogeneous GL3 luciferase expression. Experimental conditions are as in Fig. 3.
To strengthen this observation further, we measured the IFN-β production of transfected MCF-7 cells, which possess a functional IFN pathway (25). As found by Dahlgren et al. (25), 100 nM siRNA transfection led to minimal IFN-β production after 24 h (4 ± 2 pg/ml). Similar low levels of IFN-β production (4 ± 4 pg/ml) were observed after (dA)8-GL3-(dT)8 and (dT)8-GL3-(dT)8 + (dA)25 ssiRNA transfection.
siRNA Oligomerization Enhances Silencing Regardless of Aggregation or Delivery Means.
Self-aggregation by complementary sticky 3′ overhangs is the simplest way of forming larger linear nucleic acid structures with enhanced electrostatic binding to the polycationic vector. Hybridization of identical overhangs with an extra complementary single-stranded oligonucleotide or triplex-forming oligonucleotide may reach the same goal with less defined structures.
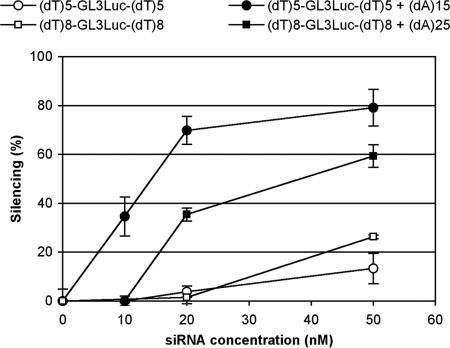
We tested such alternative solutions by mixing (dT)5-GL3Luc-(dT)5 or (dT)8-GL3Luc-(dT)8 with equimolar amounts of (dA)15 or (dA)25, respectively, before complex formation with PEI. As shown in Fig. 5, addition of the single-stranded complementary oligonucleotide led to enhancement of luciferase gene silencing. Here, the exact nature of the structure(s) involved in oligomerization is not known. The length of the single-stranded oligo was chosen such as to avoid loop formation with a single siRNA molecule. However, formation of nucleic acid networks bridged by antiparallel T·A helices and antiparallel/parallel T·A·T triple helices (26) are possible in the presence of a polycation. Other experiments showed that using either oligoribo or oligodeoxyribo sequences as added single-stranded oligonucleotides led to significant enhancement of silencing (data not shown).
Fig. 5.
Hybridization of identical (dT)n overhangs with a complementary oligodeoxyadenylate significantly enhances silencing. Experimental conditions are as in Fig. 3.
Finally, we also tested whether a cationic lipid carrier could benefit from siRNA aggregation. And indeed jetSi-ENDO-mediated silencing was found to be more efficient with aggregating siRNAs and reached a plateau at 50 nM siRNA (Fig. 6).
Fig. 6.
siRNA oligomerization enhances silencing when delivered by the cationic lipid formulation jetSi-ENDO (N/P = 6). Hybridization conditions are as in Fig. 5, and other experimental conditions are as in Fig. 3.
Taken together, a first interpretation of these results may be that ssiRNA complexes are more efficient than siRNA complexes because they better withstand competitive exchange with syndecans and other extracellular polyanions. However, results shown in Fig. 6, which were not expected after those of Fig. 1 (where lipid/siRNA complexes did enter cells), show that other parameters may come into play as well, such as the rate of siRNA release in the cytoplasm, by competition with phosphatidylserine/ethanolamine (27) in the case of jetSi-ENDO, or with polyanionic proteins (28) in the case of PEI.
Linear PEI/ssiRNA Complexes Show Enhanced Silencing in the Mouse Lung.
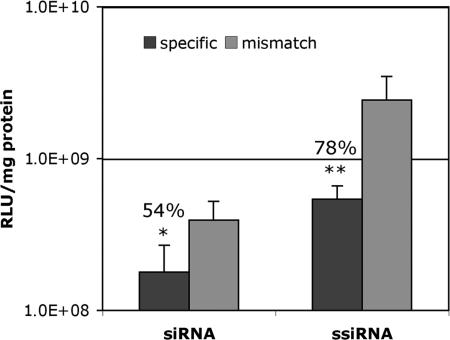
Linear PEI-mediated luciferase gene delivery and expression in the mouse is highest after i.v. injection. We previously measured higher expression levels in the lungs than in any other organ (29, 30). We therefore tested whether ssiRNA would have any advantage over siRNA in this experimental setup of transiently transfected mouse lungs. To increase the likelihood of having a lung cell receiving the luciferase gene and the silencing RNA, complexes were coinjected. Thirty micrograms of the luciferase-expressing plasmid complexed to in vivo-jetPEI (N/P = 8) and 20 μg siRNA or ssiRNA complexed to jetPEI (N/P = 8) were injected retroorbitally into OF1 mice. Lungs were removed after 24 h, and luciferase enzyme activity was measured. Results are shown in Fig. 7. The control mismatch siRNA- and ssiRNA-injected mice showed different expression levels, presumably because in vitro and in vivo transfection is enhanced by “stuffer” DNA (31). ssiRNA complexes being more stable than those of siRNA (see Fig. 2), their stuffer effect on luciferase plasmid transfection is more pronounced. As found in cell culture, ssiRNA silencing in the lung (78%) was more pronounced (P < 0.01) than silencing with the classical siRNA (54%; P < 0.05). These results are an indirect indication that concatemerization is increasing in vivo stability of PEI/ssiRNA complexes and a direct indication of the beneficial effect of stabilization on gene silencing.
Fig. 7.
Luciferase gene silencing in the mouse lung. OF1 mice were coinjected retroorbitally with a luciferase-bearing plasmid and siRNA complexed separately with jetPEI. Luciferase gene expression was measured after 24 h. ssiRNA showed significantly higher gene silencing than classical siRNA (∗, P < 0.05; ∗∗, P < 0.01).
Conclusion
Although the underlying mechanisms still require clarification, ssiRNAs are significantly more effective than siRNAs in gene silencing. The difference is not caused by faster sedimentation in cell culture or better diffusion in vivo because siRNA and ssiRNA polyplexes are both large (0.5–1 μm after 30 min) in saline and small (40–50 nm) in 5% glucose solution (results not shown). More experiments are required, with other reagents and target genes, to fully appreciate their usefulness. For gene silencing experiments in cell culture, ssiRNAs advantages may be marginal because siRNA delivery with some second-generation cationic lipids is already efficient with as little as 1 nM siRNA. At such concentrations, neither toxicity nor cost remain arguments.
The situation, however, is dramatically different in vivo, where lipid-based gene delivery has made little progress over the last decade in comparison to linear PEI. The goal of this work was to make PEI amenable to more efficient in vivo siRNA delivery. Proof of concept was demonstrated by enhanced ssiRNA-mediated luciferase silencing in transiently transfected mouse lungs. ssiRNA now awaits pertinent animal disease models.
Materials and Methods
Transfection Reagents and Nucleic Acids.
jetSi-ENDO (cationic lipid transfection reagent), jetPEI (cationic linear PEI transfection reagent), and in vivo-jetPEI were from Polyplus-Transfection. PAGE-purified nucleic acids were purchased from Eurogentec (Brussels, Belgium) and stored at −20°C as 20 μM solutions as indicated by the manufacturer. Annealing was performed in annealing buffer (Eurogentec) for 2 min at 95°C followed by slow cooling. Sequences were as follows: GL3Luc siRNA sense, 5′-CUUACGCUGAGUACUUCGATT; 5′-6-FAM-labeled anti-lamin A/C siRNA sense, 5′-CUGGACUUCCAGAAGAACATT; and vimentin siRNA sense, 5′-GAAUGGUACAAAUCCAAGU. The 21-mer dsDNA sequence used for EtdBr intercalation studies was that of the GL3Luc siRNA given above, with U replaced by T.
Confocal Microscopy.
HeLa cells (human cervix epithelial adenocarcinoma; CCL-2; ATCC, Manassas, VA) were cultured in four-well-chambered cover glasses (Lab-Tek; Nunc, Roskilde, Denmark) with 1 ml of Eagle's MEM (Eurobio, Courtaboeuf, France). Complexes were obtained by mixing siRNA (100 nM in 50 μl of 150 mM NaCl) with the transfection reagent [1.4 μl (N/P = 5) for jetPEI and 2.1 μl (N/P = 6) for jetSi-ENDO] in 50 μl of 150 mM NaCl during 30 min. Complexes were added to the cells maintained in 0.5 ml of MEM. After 4 h, 0.4 ml of MEM was added to the cells. Cells were observed live 24 h posttransfection after washing with 2× 1 ml of PBS (Eurobio) and addition of 750 μl MEM without phenol red. Images were recorded with a TCS SP2 AOBS confocal microscope (Leica, Vienna) coupled to Leica confocal software, using a ×40 oil-immersion objective and a 488-nm excitation line; confocal sections were taken every micrometer. Images were processed without loss of information by using Photoshop (Adobe Systems, San Jose, CA).
EtdBr Fluorescence Measurements.
Complex formation between double-stranded GL3Luc oligodeoxyribonucleotide and jetPEI or jetSi-ENDO, and nucleic acid exchange with chondroitin sulfate A (Sigma/Aldrich, St. Louis, MO) were studied by using a fluorimeter (Fluorolog; Horiba, Kyoto, Japan) set at 530-nm excitation and 590-nm emission wavelength.
EtdBr (Sigma/Aldrich; 4.75 μM final) and GL3Luc DNA oligonucleotide (5 μM base pairs final) were mixed in 600 μl of 5% glucose solution and fluorescence was recorded (Fi). The desired amount of transfection reagent (4 μl of jetPEI or 6 μl of jetSi-ENDO) in 300 μl of the same buffer was added to the DNA solution. After 30-min incubation, fluorescence had decreased by >97%. Chondroitin sulfate A (Sigma/Aldrich; 2.4 μl of a 2.5 mg/ml stock solution) was added to the complexes at a 4-fold anionic charge excess, and the fluorescence of the mixture was measured after 30 min (Fc). The percentage of released DNA was calculated as 100 × Fc/Fi. Preliminary experiments had shown that equilibrium was reached at the indicated times.
Stability of PEI/ssiRNA Complexes.
For complex stability studies, the required amounts of siRNA and jetPEI were each diluted in 50 μl of 150 mM NaCl. The jetPEI solution was added to the siRNA solution and vortex-mixed during 10 s, giving a 250 nM solution of siRNA complexed to jetPEI (N/P = 5). After 30-min incubation, 2 μl of loading buffer (30% glycerol/3% SDS/0.4% bromophenol blue in 20 mM Tris·HCl, pH 7.8) was added to a 15-μl sample of complexes and loaded on a 1.2% NuSieve GTG agarose gel (Cambrex, East Rutherford, NJ) in 1× TAE solution (Eurobio). For HS challenge, after 30-min complex formation, siRNA/PEI complexes were incubated for 30 min at room temperature with the desired amount of HS (Sigma/Aldrich). Fifteen-microliter samples of siRNA/PEI/HS solution were mixed with 2 μl of loading buffer and loaded on a 1.2% agarose gel. siRNA was visualized under UV illumination after 15 min of soaking in a 0.5 μg/ml EtdBr solution.
For chemical stability studies, 50 μg of GL3Luc siRNA or (dA)8-GL3Luc-(dT)8 ssiRNA was complexed with jetPEI (N/P = 8) in 0.2 ml 5% glucose. Aliquots corresponding to 0.5 μg of siRNA or ssiRNA were incubated for 30 min at 37°C with increasing amounts (0, 0.1, 0.5, 1, 5, 10, 50, and 100 μg) of RNase A (Promega, Charbonnieres, France) in RNase digestion buffer in a total volume of 20 μl. The samples were incubated with 0.65% SDS and 7 μg of proteinase K (Sigma/Aldrich) for 30 min at 37°C and subjected to electrophoresis on a 1.5% agarose gel in 1× TAE solution (Eurobio) at 60 V for 2 h. Intact RNA was visualized under UV illumination after 15 min of EtdBr soaking. Band migrations and intensities were reported vs. controls without RNase and quantified by densitometry by using SynGene gene tools software.
Cell Culture.
A549 cells (human lung carcinoma; CCL-185; ATCC) stably expressing GL3 luciferase (P. pyralis luciferase under control of simian virus 40 elements) were obtained after stable transfection of pGL3Luc plasmid (Clontech, Mountain View, CA). A549Luc cells were grown in RPMI medium 1640 (Eurobio), supplemented with 10% FBS (Perbio, Brebieres, France), 2 mM glutamax (Eurobio), 100 units/ml penicillin (Eurobio), 100 μg/ml streptomycin (Eurobio), and 0.8 μg/ml G418 (Promega). Cells were maintained at 37°C in a 5% CO2 humidified atmosphere.
Transfection Experiments.
The day before transfection, 2.5 × 104 cells were seeded in 24-well tissue culture plates in 1 ml of fresh complete medium containing 10% FBS. Complexes were prepared as follows: for a triplicate experiment, the required amounts of siRNA and transfection reagent were separately diluted in 150 μl of serum-free medium for jetSi-ENDO or 150 μl of NaCl 150 mM for jetPEI. Three and 2 μl of jetSi-ENDO (N/P = 6) and jetPEI (N/P = 5) was used per μg of siRNA, respectively. For experiments with oligodeoxyadenylates, the oligonucleotide and siRNA were mixed before complex formation, and the N/P included the total amount of nucleic acid. Solutions were vortex-mixed for 10 s and left for 10 min at room temperature. The transfection reagent was added to the siRNA solution, vortex-mixed for 10 s, and left 30 min at room temperature. The complete medium was removed from the cells and replaced by 0.5 ml of serum-free medium. Complexes (100 μl per well) were added to the cells, and the plates were hand-rotated to ensure homogeneity and incubated at 37°C. After 2-h incubation, the medium was removed and replaced by 1 ml of complete medium containing 10% serum. The plate was further incubated at 37°C for 48 h.
Luciferase and Protein Assays.
Luciferase gene expression was measured with a commercial kit (Promega). After removing the cell culture medium, cells were rinsed with 1 ml of PBS. Cells were lysed with 100 μl of lysis buffer (Promega) per well, and the plate was left at room temperature for 30 min. The lysates were collected and centrifuged at 6,000 × g for 5 min. Luciferase enzyme activity was assessed by using 5 μl of lysate after addition of 100 μl of luciferin solution. The luminescence (expressed as RLU) was integrated over 10 s with a luminometer (Centro LB 960; Berthold, Thoiry, France). Luciferase activity was expressed as light units integrated over 10 s (RLU) and normalized per mg of cell protein by using the BCA assay (Pierce, Brebieres, France). Experiments were done in triplicate, and luciferase silencing efficiency was calculated relative to the luciferase level of nontransfected A549Luc cells. Negative controls were performed for each siRNA concentration with GL2Luc siRNA.
Vimentin Silencing.
NIH 3T3 cells (mouse embryonic fibroblast, CRL-1658) were grown in DMEM (Eurobio) supplemented with 4 mM l-glutamine, 1.5 g/liter sodium bicarbonate, 4.5 g/liter glucose, antibiotics (penicilline/streptomycine), and 10% FBS.
The cells (5 × 104 cells per well, 24-well plate) were transfected with 100, 75, and 50 nM normal and A8·T8 sticky vimentin siRNA complexed with jetPEI (N/P = 8) as described. GL2Luc siRNA and A8·T8 ssiRNA were used as negative controls. After 72 h, the cells were washed with 1 ml of PBS (Cambrex) and trypsinized with 100 μl trypsin/EDTA (Euromedex, Souffelweyersheim, France). Addition of 0.5 ml of complete medium containing 10% serum stopped trypsin. The wells were pooled per triplicate and centrifuged, and the pellet was washed with PBS. After centrifugation, the pellet was lysed in 100 μl of RIPA buffer (50 mM Tris·HCl, pH 7.4/150 mM NaCl/1 mM EDTA/1% Triton X-100/1% sodium deoxycholate/0.1% SDS) for 20 min at 4°C. The lysate was homogenized with a vortex-mixer and centrifuged for 5 min at 6,000 × g. Total protein content was assessed with the BCA kit (Pierce). Volumes corresponding to 1 μg of total protein were subjected to electrophoresis on a 10% acrylamide/bisacrylamide gel and transferred to a poly(vinylidene difluoride) membrane (Millipore, Molsheim, France). Vimentin expression was detected with a guinea pig anti-vimentin polyclonal antibody (RDI, Flanders, NJ) diluted at 1/3,000. GAPDH was detected with a mouse anti-GAPDH monoclonal antibody (Ambion, Austin, TX) and was used to normalize the protein level. The immunoreactive proteins were visualized by using horseradish peroxidase-conjugated anti-guinea pig or anti-mouse antibodies and an Amplified Opti-4CN Substrate Kit (Bio-Rad, Hercules, CA) using the manufacturer's instructions. Bands were quantified by densitometry using SynGene gene tools software.
IFN-β Production.
MCF-7 cells (ATCC, HTB-22; 5 × 104 cells per well in 24-well tissue culture plates) were transfected with jetPEI polyplexes formed with100 nM GL3Luc siRNA, (dA)8-GL3Luc-(dT)8 ssiRNA, or (dT)8-GL3Luc-(dT)8 + (dA)25 ssiRNA as described. After 24 h the supernatants were collected for cytokine measurement. The amount of IFN-β was determined on 100 μl of supernatant, loaded in duplicate, by sandwich ELISA (BioSource International, Camarillo, CA). Transfection of 100 nM poly(I):poly(C) (Sigma/Aldrich) was taken as positive control (18 ± 0.8 pg IFN-β/ml).
Animal Experiments.
Thirty micrograms of pGL2Luc and 20 μg of siRNA, each in 50 μl of 5% glucose, were mixed separately with in vivo-jetPEI to a N/P of 8. The final volume of each complex was 100 μl. The GL2 (interfering) and GL3 (mismatch control) siRNA sequences are given in Results. The cognate ssiRNAs had 3′ (dA)8 and (dT)8 overhangs, hence their molar concentrations were 40% lower than the siRNAs. After 15-min incubation the plasmid complexes and siRNA complexes were mixed together. A total of 200 μl of DNA/siRNA mixture was injected retroorbitally into 8-week-old OF1 female mice (Charles River Laboratory, L'Arbresle, France). After 24 h, mice (n = 4) were anesthetized by i.p. injection of pentobarbital (40 mg/kg; Ceva, Libourne, France), the lung was dissected, rinsed with PBS, and mixed with an ultra-thurax homogenizer in 2 ml of 1× lysis buffer (Promega). Each organ mix was frozen at −80°C and thawed, and an aliquot of 0.5 ml was taken for luciferase analysis. The aliquot was centrifuged for 5 min at 14,000 × g. Luciferase enzyme activity was assessed on 5 μl of organ supernatant lysate using 100 μl of luciferin solution (Promega). The luminescence (expressed as RLU) was integrated over 10 s by using a luminometer (Centro LB960; Berthold). Luciferase activity was expressed as light units integrated over 10 s and normalized per mg of organ protein with the BCA assay (Pierce). For statistical analysis of differences between cognate and mismatch sequences an unpaired Student's t test was used.
Abbreviations
- PEI
polyethylenimine
- ssiRNA
siRNA with short sticky overhang
- EtdBr
ethidium bromide
- N/P
PEI amine over DNA phosphate charge ratio
- HS
heparan sulfate
- RLU
relative light units.
Footnotes
The authors declare no conflict of interest.
References
- 1.Morris KV, Rossi JJ. Curr Opin Mol Ther. 2006;8:115–121. [PubMed] [Google Scholar]
- 2.Manoharan M. Curr Opin Chem Biol. 2004;8:570–579. doi: 10.1016/j.cbpa.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 3.Boussif O, Lezoualc'h F, Zanta MA, Mergny MD, Scherman D, Demeneix B, Behr JP. Proc Natl Acad Sci USA. 1995;92:7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aigner A. J Biotechnol. 2006;124:12–25. doi: 10.1016/j.jbiotec.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 5.Goula D, Remy JS, Erbacher P, Wasowicz M, Levi G, Abdallah B, Demeneix BA. Gene Ther. 1998;5:712–717. doi: 10.1038/sj.gt.3300635. [DOI] [PubMed] [Google Scholar]
- 6.Kawakami S, Ito Y, Charoensit P, Yamashita F, Hashida M. J Pharmacol Exp Ther. 2006;317:1382–1390. doi: 10.1124/jpet.105.100669. [DOI] [PubMed] [Google Scholar]
- 7.Merdan T, Kunath K, Fischer D, Kopecek J, Kissel T. Pharm Res. 2002;19:140–146. doi: 10.1023/a:1014212630566. [DOI] [PubMed] [Google Scholar]
- 8.Akinc A, Thomas M, Klibanov AM, Langer R. J Gene Med. 2005;7:657–663. doi: 10.1002/jgm.696. [DOI] [PubMed] [Google Scholar]
- 9.Zabner J, Fasbender AJ, Moninger T, Poellinger KA, Welsh MJ. J Biol Chem. 1995;270:18997–19007. doi: 10.1074/jbc.270.32.18997. [DOI] [PubMed] [Google Scholar]
- 10.Kopatz I, Remy JS, Behr JP. J Gene Med. 2004;6:769–776. doi: 10.1002/jgm.558. [DOI] [PubMed] [Google Scholar]
- 11.Kokenyesi R, Bernfield M. J Biol Chem. 1994;269:12304–12309. [PubMed] [Google Scholar]
- 12.Sanderson RD, Turnbull JE, Gallagher JT, Lander AD. J Biol Chem. 1994;269:13100–13106. [PubMed] [Google Scholar]
- 13.Tumova S, Woods A, Couchman JR. Int J Biochem Cell Biol. 2000;32:269–288. doi: 10.1016/s1357-2725(99)00116-8. [DOI] [PubMed] [Google Scholar]
- 14.Radler JO, Koltover I, Salditt T, Safinya CR. Science. 1997;275:810–814. doi: 10.1126/science.275.5301.810. [DOI] [PubMed] [Google Scholar]
- 15.Naqvi KR, Behr J-P, Chapman D. Chem Phys Lett. 1974;26:440–444. [Google Scholar]
- 16.Ruponen M, Yla-Herttuala S, Urtti A. Biochim Biophys Acta. 1999;1415:331–341. doi: 10.1016/s0005-2736(98)00199-0. [DOI] [PubMed] [Google Scholar]
- 17.Pörschke D. Biopolymers. 1971;10:1989–2013. [Google Scholar]
- 18.Higuchi S, Tsuboi M. Biopolymers. 1966;4:837–854. doi: 10.1002/bip.1966.360040804. [DOI] [PubMed] [Google Scholar]
- 19.Behr JP. Tetrahedron Lett. 1986;27:5861–5864. [Google Scholar]
- 20.Pontius BW, Berg P. Proc Natl Acad Sci USA. 1991;88:8237–8241. doi: 10.1073/pnas.88.18.8237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mao S, Neu M, Germershaus O, Merkel O, Sitterberg J, Bakowsky U, Kissel T. Bioconjugate Chem. 2006;17:1209–1218. doi: 10.1021/bc060129j. [DOI] [PubMed] [Google Scholar]
- 22.Thompson JF, Hayes LS, Lloyd DB. Gene. 1991;103:171–177. doi: 10.1016/0378-1119(91)90270-l. [DOI] [PubMed] [Google Scholar]
- 23.Kafri T, van Praag H, Gage FH, Verma IM. Mol Ther. 2000;1:516–521. doi: 10.1006/mthe.2000.0083. [DOI] [PubMed] [Google Scholar]
- 24.Elbashir SM, Harborth J, Weber K, Tuschl T. Methods. 2002;26:199–213. doi: 10.1016/S1046-2023(02)00023-3. [DOI] [PubMed] [Google Scholar]
- 25.Dahlgren C, Wahlestedt C, Thonberg H. Biochem Biophys Res Commun. 2006;341:1211–1217. doi: 10.1016/j.bbrc.2006.01.085. [DOI] [PubMed] [Google Scholar]
- 26.Torigoe H, Ferdous A, Watanabe H, Akaike T, Maruyama A. J Biol Chem. 1999;274:6161–6167. doi: 10.1074/jbc.274.10.6161. [DOI] [PubMed] [Google Scholar]
- 27.Berezhna S, Schaefer S, Heintzmann R, Jahnz M, Boese G, Deniz A, Schwille P. Biochim Biophys Acta. 2005;1669:193–207. doi: 10.1016/j.bbamem.2005.02.011. [DOI] [PubMed] [Google Scholar]
- 28.Okuda T, Niidome T, Aoyagi H. J Control Release. 2004;98:325–332. doi: 10.1016/j.jconrel.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 29.Zou SM, Erbacher P, Remy JS, Behr JP. J Gene Med. 2000;2:128–134. doi: 10.1002/(SICI)1521-2254(200003/04)2:2<128::AID-JGM95>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 30.Goula D, Becker N, Lemkine GF, Normandie P, Rodrigues J, Mantero S, Levi G, Demeneix BA. Gene Ther. 2000;7:499–504. doi: 10.1038/sj.gt.3301113. [DOI] [PubMed] [Google Scholar]
- 31.Kichler A, Leborgne C, Danos O. J Gene Med. 2005;7:1459–1467. doi: 10.1002/jgm.805. [DOI] [PubMed] [Google Scholar]