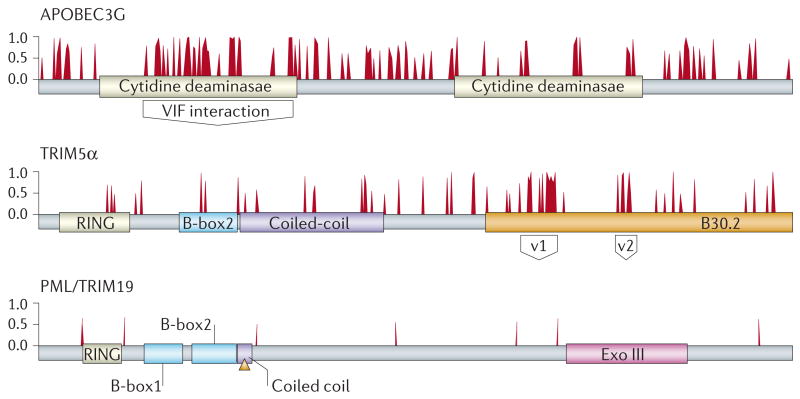
Figure 2. Comparative and evolutionary analysis of host proteins involved in HIV-1 pathogenesis.
This analysis used a comprehensive set of sequences that are representative of the primate lineages — new world monkeys, old world monkeys, gibbons and apes — to identify signs of positive and of negative selection in protein domains. The three proteins depicted represent different patterns of evolution in primates. The antiretroviral protein APOBEC3G is characterized by many residues that are under positive selective pressure, with a large cluster that delineates the virion infectivity factor (Vif)-interaction domain. These amino acids contribute to discrimination among the various Vif proteins of HIV and different SIVs. A second antiretroviral protein, TRIM5α (tripartite motif 5α) has a patch of positively selected residues that defines the variable 1 region (v1) and variable 2 region (v2) and that carry the capsid recognition specificity. The variable regions of TRIM5α might have evolved independently to recognize various retroviruses. By contrast, there are no residues under positive selective pressure in the PML/TRIM19 protein, which is also proposed to have antiviral properties. Therefore, the effect of PML/TRIM19 might be indirect. Failure to identify a signature of positive selection militates against a more direct role for this protein in antiviral defence, because it would be expected that prolonged contact with several pathogens over long evolutionary time periods would have resulted in signatures of positive selection indicative of a genetic conflict. Bars represent amino acids that are predicted to be under positive selection in a grading of posterior probability from 0 to 1. The various protein domains are indicated. Adapted with permission from Ref. 31.