Fig. 5.
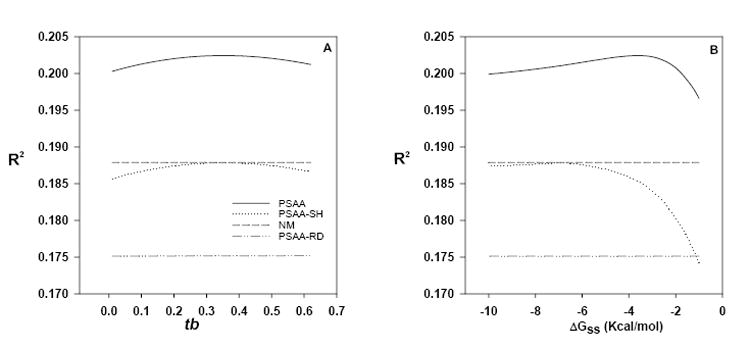
Effects of changing tb or ΔGss-cutoff on the performance of the PSAA model. Effects of changing (A) the values of tb while holding ΔGss-cutoff = -3.6 or (B) the values of ΔGss-cutoff while holding tb = 0.35 on the performance of PSAA: the position-dependent secondary-structure attenuated affinity model (Eq. 6). Data shown are for the human genome U133 Latin Square Experiment 2 Replicate 1 PM probes. NM: Naef and Magnasco (Naef and Magnasco, 2003) model (Eq. 5). The suffixes (-SH) and (-RD) indicates the R2 after generating the minimum folding energy (ΔGss) and the minimum energy structure from shuffled and random sequences, respectively (see section 3.3 for explanation).
