Abstract
Tigecycline, a member of the glycylcycline class of antibiotics, was designed to maintain the antibacterial spectrum of the tetracyclines while overcoming the classic mechanisms of tetracycline resistance. The current study was designed to monitor the prevalence of the tet(A), tet(B), tet(C), tet(D), tet(E), and tet(M) resistance determinants in Escherichia coli isolates collected during the worldwide tigecycline phase 3 clinical trials. A subset of strains were also screened for the tet(G), tet(K), tet(L), and tet(Y) genes. Of the 1,680 E. coli clinical isolates screened for resistance to classical tetracyclines, 405 (24%) were minocycline resistant (MIC ≥ 8 μg/ml) and 248 (15%) were tetracycline resistant (MIC ≥ 8 μg/ml) but susceptible to minocycline (MIC ≤ 4 μg/ml). A total of 452 tetracycline-resistant, nonduplicate isolates were positive by PCR for at least one of the six tetracycline resistance determinants examined. Over half of the isolates encoding a single determinant were positive for tet(A) (26%) or tet(B) (32%) with tet(C), tet(D), tet(E), and tet(M), collectively, found in 4% of isolates. Approximately 33% of the isolates were positive for more than one resistance determinant, with the tet(B) plus tet(E) combination the most highly represented, found in 11% of isolates. The susceptibilities of the tetracycline-resistant strains to tigecycline (MIC90, 0.5 μg/ml), regardless of the encoded tet determinant(s), were comparable to the tigecycline susceptibility of tetracycline-susceptible strains (MIC90, 0.5 μg/ml). The results provide a current (2002 to 2006) picture of the distribution of common tetracycline resistance determinants encoded in a globally sourced collection of clinical E. coli strains.
Escherichia coli, the causative agent of a number of infections, such as gastroenteritis and cystitis in nonhospitalized patients and pneumonia and septicemia of mostly nosocomial origin (13), has acquired resistance to many antibiotics, including the tetracycline class of agents (1, 7, 10). Widespread resistance to the broad-spectrum tetracyclines has been caused, in part, by heavy clinical use and misuse in the human population. Additional contributing factors are the use of tetracyclines in agriculture as a growth promoter and as an infection control agent in domestic animals, aquaculture, and horticulture (9, 10, 24). In the United States and other parts of the world, tetracyclines, alone or in combination with other antibiotics, are still used for the treatment of human infections, as well as for prophylaxis, both orally and topically, due to their excellent safety profile and low cost (32).
Tetracycline resistance in bacteria is mediated by four mechanisms: efflux, ribosomal protection, enzymatic inactivation, and target modification (10). Tetracycline efflux, first identified in 1953 in Shigella dysenteriae (1), and ribosomal protection, first identified in Streptococcus spp. (7), are now prevalent in both gram-negative and gram-positive bacteria (10). These resistance mechanisms are widely distributed in bacteria due to their association with mobilizable DNA elements, which have facilitated the spread to more than 50 genera, and are often coupled with multidrug resistance (11, 31, 33). Tetracycline resistance mediated through enzymatic inactivation and target modification has only been identified in a few bacterial species (34, 42) and, at present, is of limited clinical importance.
At present, 23 genes encoding efflux pumps and 11 genes encoding ribosomal protection proteins, not including the recently described mosaic tetracycline resistance genes (27), have been identified in bacteria since the first report of transferable tetracycline resistance in 1960 (33). The leading tetracycline resistance mechanism in E. coli is the extrusion of drug from the cytoplasm via efflux (10). Tetracycline-specific efflux pumps are members of the major facilitator superfamily (MFS) of efflux pumps (28). MFS pumps specific for tetracycline operate by transporting tetracycline in an energy-dependent fashion, via proton exchange, thereby reducing the intracellular concentration of the drug (10). Tet pumps are divided into six groups based on amino acid sequence, with Tet(A), Tet(B), Tet(C), Tet(D), and Tet(E) placed in group 1 due to amino acid sequence similarity (10, 33). Most tetracycline-specific efflux pumps confer resistance to tetracycline only; however, tet(B) encodes a pump that is able to extrude both tetracycline and minocycline (18, 29).
Tigecycline is the novel 9-t-butyl glycylamido derivative of minocycline that has been approved for use in complicated skin and skin structure infections and complicated intra-abdominal infections (2, 14) (Tygacil package insert, http://www.fda.gov/cder/foi/label/2005/021821lbl.pdf; Wyeth Pharmaceuticals Inc., Collegeville, PA). During the course of the tigecycline phase 3 clinical trials, all bacterial isolates were screened for susceptibility to tetracycline, among a panel of antibiotics. Tetracycline-resistant E. coli isolates (MIC ≥ 8 μg/ml) were examined by PCR for the presence of 10 resistance determinants: tet(A), tet(B), tet(C), tet(D), tet(E), tet(G), tet(K), tet(L), tet(M), and tet(Y).
MATERIALS AND METHODS
Strains.
The E. coli clinical isolates were collected from patients enrolled worldwide in the tigecycline clinical trials, and pure cultures of each isolate were submitted to a central laboratory. Identification was performed by using a Vitek system (BioMerieux, Durham, NC). The bacterial isolates were considered to be pathogens by the principal investigator for the study and came from all patients regardless of treatment with tigecycline or comparator. The strains were riboprinted using a RiboPrinter microbial characterization system (Qualicom, Wilmington, DE) according to the manufacturer's instructions. Only one isolate per patient was included in the study unless ribotyping determined that serial isolates collected from a patient were unique (nonduplicate) strain types.
Susceptibility determination.
Broth microdilutions were conducted in accordance with Clinical and Laboratory Standards Institute recommendations, using fresh Mueller Hinton II broth (MHB) for tigecycline (12).
Primers, template preparation, and PCR assay.
The primer sets, PCR assay conditions, and positive control strains known to harbor the specific determinants for each primer set were previously described (22). In addition, for the present work, PCR assays were designed to detect the tet(G) (GenBank accession no. AF133139), tet(Y) (GenBank accession no. AB089608), and tet(L) (GenBank accession no. U17153) genes. The primer set used for the amplification of tet(G) was F, 5′CAT TGC CCT GCT GAT CG; and R, 5′ TTG GTG AGG CTT GTA AGC. The following primers were used for tet(Y): F, 5′ CCG CAC TCA TTG TTG TCG; R, 5′ TTT TCA TCG CAA ACA AGA CC. The primer set used for the detection of tet(L) was F, 5′ ATA AAT TGT TTC GGG TCG GTA AT; and R, 5′ AAC CAG CCA ACT AAT GAC AAT (8). The primers and conditions for tet(K) (GenBank accession no. AJ888003) amplification were previously described (21). The positive control strains were Salmonella enterica serovar Typhimurium strain DT104 (5, 37) for tet(G) and Enterococcus faecalis strain BM4253 for tet(L) (8) and a recombinant E. coli DH5α strain transformed with plasmid pIE1120 containing tet(Y), kindly provided by E. Tietze (38, 39). The expected size of the amplification product for each target gene was as follows: tet(G), 993 bp; tet(K), 1.08 kb; tet(L), 1.07 kb; and tet(Y), 949 bp. Lysate production and PCR amplifications were carried out as previously described (22).
RESULTS
Resistance determinant identification.
A total of 1,680 E. coli clinical isolates collected during the tigecycline phase 3 clinical trials for complicated skin and skin structure infections, complicated intra-abdominal infections, community-acquired pneumonia, and hospital-acquired pneumonia were screened for tetracycline resistance. A total of 405 (24%) of the strains were resistant to both minocycline and tetracycline (MIC ≥ 8 μg/ml), and 248 (15%) were tetracycline resistant but susceptible to minocycline (MIC ≤ 4 μg/ml). The 653 resistant isolates were screened by PCR for the common tetracycline determinants tet(A), tet(B), tet(C) tet(D), tet(E), and tet(M). Ribotyping determined that 488 isolates were nonduplicates on the patient level (data not shown). Of these, 452 (93%) were positive for one or more of the six tested determinants, whereas 36 (7%) isolates were negative for all six tetracycline resistance determinants on the original screening panel. These 36 isolates were further screened for the presence of the tet(G) and tet(Y) genes, which have been previously described in E. coli (5, 39), as well as the tet(K) and tet(L) genes, which have been detected in some gram-negative bacteria (33). All 36 isolates were negative for tet(G), tet(K), tet(L), and tet(Y) (data not shown); the basis of tetracycline resistance in these isolates is the focus of ongoing research.
Worldwide prevalence of tetracycline resistance determinants.
An examination of single tetracycline resistance determinants revealed that tet(A) and tet(B) accounted for 58% of all isolates, with 26% of isolates encoding only tet(A) and 32% of isolates encoding only tet(B) (Table 1). The other efflux pump-encoding genes were found much less frequently, with 3% of isolates encoding tet(D), 1% of isolates encoding tet(C), and only a single strain encoding the tet(E) gene. Interestingly, the tet(E) determinant in combination with tet(B) (11%) was found more frequently than the tet(E) determinant alone (0.2%); in fact this was the most common combination of multiple efflux determinants identified (Table 2). The other two most frequent combinations were the tet(A) plus tet(D) pair and the tet(B) plus tet(D) pair seen in 5.1% and 6.6% of isolates, respectively. Overall, 33% of isolates encoded more than one tetracycline resistance determinant, with 2.5% of isolates encoding three or more determinants.
TABLE 1.
Regional distribution of E. coli isolates with a single tetracycline resistance determinant
| Determinant | No. (%) of isolates with determinant froma:
|
Totalb | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Asia-Pacific | Australia | Eastern Europe | India | Latin America | North America | South Africa | Western Europe | ||
| tet(A) | 19 (27.9) | 1 (33.3) | 42 (29) | 4 (11.4) | 24 (19.7) | 9 (19.6) | 9 (40.9) | 17 (37) | 125 (25.6) |
| tet(B) | 18 (26.5) | 1 (33.3) | 49 (33.5) | 12 (34.3) | 50 (41) | 15 (32.6) | 6 (27.3) | 6 (12.8) | 157 (32.2) |
| tet(C) | 2 (5.7) | 1 (0.8) | 2 (4.3) | 5 (1) | |||||
| tet(D) | 6 (8.8) | 2 (1.4) | 1 (2.9) | 3 (2.5) | 1 (2.2) | 13 (2.7) | |||
| tet(E) | 1 (0.7) | 1 (0.2) | |||||||
| tet(M) | 1 (0.8) | 1 (0.2) | |||||||
| Totalc | 68 | 3 | 146 | 35 | 122 | 46 | 22 | 46 | 302 |
Asia-Pacific: China, Korea, Taiwan. Eastern Europe: Bulgaria, Czech Republic, Estonia, Herzegovina, Hungary, Latvia, Lithuania, Poland, Romania, Russia, Slovakia, Ukraine. Latin America: Argentina, Brazil, Chile, Guatemala, Mexico, Panama, Peru. North America: United States, Canada. Western Europe: Austria, Belgium, France, Germany, Great Britain, Italy, Spain.
Total number of isolates with the given tetracycline resistance determinant collected from all sampled regions.
Total number of tetracycline-resistant isolates collected from each region.
TABLE 2.
Regional distribution of E. coli isolates with multiple tetracycline resistance determinants
| Determinants | No. (%) of isolates with determinants froma:
|
Totalb | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Asia-Pacific | Australia | Eastern Europe | India | Latin America | North America | South Africa | Western Europe | ||
| tet(A), tet(B) | 3 (4.4) | 3 (2) | 3 (8.6) | 5 (4.1) | 2 (4.3) | 1 (2.1) | 17 (3.5) | ||
| tet(A), tet(D) | 7 (10.3) | 10 (6.8) | 2 (1.6) | 1 (4.5) | 5 (10.6) | 25 (5.1) | |||
| tet(A), tet(E) | 2 (5.7) | 2 (0.4) | |||||||
| tet(A), tet(M) | 3 (4.4) | 1 (0.7) | 2 (4.3) | 6 (1.2) | |||||
| tet(B), tet(C) | 1 (1.5) | 1 (0.2) | |||||||
| tet(B), tet(D) | 2 (2.9) | 6 (4.1) | 1 (2.9) | 10 (8.2) | 8 (17.4) | 1 (4.5) | 4 (8.5) | 32 (6.6) | |
| tet(B), tet(E) | 4 (5.9) | 1 (33.3) | 23 (15.6) | 1 (2.9) | 9 (7.4) | 3 (6.5) | 4 (18.2) | 8 (17) | 53 (10.8) |
| tet(C), tet(E) | 1 (0.8) | 1 (0.2) | |||||||
| tet(D), tet(E) | 1 (1.5) | 1 (0.2) | |||||||
| tet(A), tet(B), tet(D) | 1 (0.7) | 1 (2.9) | 1 (2.2) | 3 (0.6) | |||||
| tet(B), tet(D), tet(E) | 1 (1.5) | 4 (2.7) | 1 (0.8 | 1 (4.5) | 1 (2.1) | 8 (1.6) | |||
| tet(A), tet(B), tet(D), tet(E) | 1 (0.8) | 1 (0.2) | |||||||
| No tet marker identified | 3 (4.4) | 4 (2.7) | 8 (22.9) | 14 (11.5) | 5 (10.9) | 2 (4.3) | 36 (7.4) | ||
| Totalc | 68 | 3 | 146 | 35 | 122 | 46 | 22 | 46 | 186 |
Asia-Pacific: China, Korea, Taiwan. Eastern Europe: Bulgaria, Czech Republic, Estonia, Herzegovina, Hungary, Latvia, Lithuania, Poland, Romania, Russia, Slovakia, Ukraine. Latin America: Argentina, Brazil, Chile, Guatemala, Mexico, Panama, Peru. North America: United States, Canada. Western Europe: Austria, Belgium, France, Germany, Great Britain, Italy, Spain.
Total number of isolates with the given tetracycline resistance determinant collected from all sampled regions.
Total number of tetracycline-resistant isolates collected from each region.
With the exception of Western Europe, where the numbers of single- and multiple-determinant-encoding isolates were nearly equivalent, strains from other regions showed a trend (>55% of isolates) towards single determinants (Tables 1 and 2). In looking at single determinants, the tet(B) gene was seen in nearly 30% of isolates tested from India, Latin America, and North America, whereas in Asia-Pacific and Eastern Europe, tet(A) and tet(B) were equivalent in occurrence, and in South Africa and Western Europe, the tet(A) determinant predominated.
Multiple-determinant isolates encoding tet(B) plus tet(E) accounted for >16% of isolates from Europe (Eastern and Western) and South Africa. Similarly, multiple-determinant isolates from North America encoding the tet(B) plus tet(D) pairing accounted for 17% of isolates from that region and isolates encoding the tet(A) plus tet(D) pair accounted for 10% of isolates from the Asia-Pacific region and Western Europe (Table 2).
The study also identified seven isolates, two from Taiwan and one each from Belgium, China, France, Guatemala, and Romania, expressing tet(M) homologues encoding a ribosomal protection protein. Although reported previously in commensal E. coli isolates (25), tet(M) had not been reported in human clinical E. coli isolates until 2006 (22). All of the isolates, with the exception of the isolate from Guatemala, also encoded the tet(A) resistance determinant.
Antibiotic susceptibility.
Tetracycline and minocycline susceptibility data for the strain collection are presented in Table 3 and Fig. 1. The tet(B) gene was detected in 272 (56%) isolates, either alone or in combination with additional determinants. The minocycline susceptibility pattern of this group of strains (MIC50, 8 μg/ml; MIC90, 32 μg/ml; MIC range, 0.25 to >64 μg/ml) reflects the unique ability of TetB to efflux minocycline (18, 29). The remainder of the strains, encoding tet(A), tet(C), tet(D), tet(E), or tet(M) or a combination of these determinants, were highly resistant to tetracycline (MIC90, ≥64 μg/ml) and would be categorized as intermediate with respect to minocycline (MIC90, 8 μg/ml).
TABLE 3.
Tigecycline, minocycline, and tetracycline susceptibilities of E. coli isolates expressing various tetracycline resistance determinants
| Tetracycline resistance determinant status or strain | No. of isolates with determinant | Antibiotic | MIC (μg/ml)a
|
||
|---|---|---|---|---|---|
| Range | MIC50 | MIC90 | |||
| tet(A) | 125 | Tigecycline | 0.12-2 | 0.5 | 0.5 |
| Minocycline | 0.5-32 | 2 | 4 | ||
| Tetracycline | 8->64 | >64 | >64 | ||
| tet(B) | 157 | Tigecycline | 0.12-1 | 0.25 | 0.5 |
| Minocycline | 2->64 | 16 | 32 | ||
| Tetracycline | 8->64 | >64 | >64 | ||
| tet(C) | 5 | Tigecycline | 0.12-0.5 | NA | NA |
| Minocycline | 1-4 | NA | NA | ||
| Tetracycline | 16->64 | NA | NA | ||
| tet(D) | 13 | Tigecycline | 0.25-0.5 | 0.25 | 0.5 |
| Minocycline | 2-16 | 8 | 8 | ||
| Tetracycline | 64->64 | >64 | >64 | ||
| tet(E) | 1 | Tigecycline | 0.5 | NA | NA |
| Minocycline | 8 | NA | NA | ||
| Tetracycline | >64 | NA | NA | ||
| tet(M) | 1 | Tigecycline | 0.5 | NA | NA |
| Minocycline | 2 | NA | NA | ||
| Tetracycline | 8 | NA | NA | ||
| tet(A), tet(B) | 17 | Tigecycline | 0.25-0.5 | 0.25 | 0.5 |
| Minocycline | 4-32 | 16 | 32 | ||
| Tetracycline | 32->64 | >64 | >64 | ||
| tet(A), tet(D) | 25 | Tigecycline | 0.25-2 | 0.5 | 1 |
| Minocycline | 0.25-64 | 4 | 16 | ||
| Tetracycline | 32->64 | >64 | >64 | ||
| tet(A), tet(E) | 2 | Tigecycline | 0.5 | NA | NA |
| Minocycline | 2-4 | NA | NA | ||
| Tetracycline | >64 | NA | NA | ||
| tet(A), tet(M) | 6 | Tigecycline | 0.25-0.5 | NA | NA |
| Minocycline | 4-32 | NA | NA | ||
| Tetracycline | >64 | NA | NA | ||
| tet(B), tet(C) | 1 | Tigecycline | 0.5 | NA | NA |
| Minocycline | 64 | NA | NA | ||
| Tetracycline | 64 | NA | NA | ||
| tet(B), tet(D) | 32 | Tigecycline | 0.12-0.5 | 0.25 | 0.5 |
| Minocycline | 8->64 | 16 | 64 | ||
| Tetracycline | 8->64 | >64 | >64 | ||
| tet(B), tet(E) | 53 | Tigecycline | 0.06-0.5 | 0.25 | 0.25 |
| Minocycline | 1-64 | 8 | 32 | ||
| Tetracycline | 8->64 | >64 | >64 | ||
| tet(C), tet(E) | 1 | Tigecycline | 1 | NA | NA |
| Minocycline | 4 | NA | NA | ||
| Tetracycline | >64 | NA | NA | ||
| tet(D), tet(E) | 1 | Tigecycline | 0.25 | NA | NA |
| Minocycline | 8 | NA | NA | ||
| Tetracycline | 64 | NA | NA | ||
| tet(A), tet(B), tet(D) | 3 | Tigecycline | 0.25 | NA | NA |
| Minocycline | 8-32 | NA | NA | ||
| Tetracycline | >64 | NA | NA | ||
| tet(B), tet(D), tet(E) | 8 | Tigecycline | 0.12-0.5 | NA | NA |
| Minocycline | 8-32 | NA | NA | ||
| Tetracycline | 32->64 | NA | NA | ||
| tet(A), tet(B), tet(D), tet(E) | 1 | Tigecycline | 0.25 | NA | NA |
| Minocycline | 8 | NA | NA | ||
| Tetracycline | >64 | NA | NA | ||
| Tet-resistant isolates, no determinant identified | 36 | Tigecycline | 0.25-2 | 0.5 | 1 |
| Minocycline | 0.5-32 | 4 | 8 | ||
| Tetracycline | 8->64 | >64 | >64 | ||
| Isolates with tet(B) | 272 | Tigecycline | 0.06-1 | 0.25 | 0.5 |
| Minocycline | 0.25->64 | 8 | 32 | ||
| Tetracycline | 8->64 | >64 | >64 | ||
| Isolates without tet(B) | 180 | Tigecycline | 0.12-2 | 0.5 | 0.5 |
| Minocycline | 0.25-64 | 4 | 8 | ||
| Tetracycline | 8->64 | >64 | >64 | ||
| Single-determinant isolates | 302 | Tigecycline | 0.12-2 | 0.25 | 0.5 |
| Minocycline | 0.5->64 | 8 | 32 | ||
| Tetracycline | 8->64 | >64 | >64 | ||
| Multiple-determinant isolates | 150 | Tigecycline | 0.06-2 | 0.25 | 0.5 |
| Minocycline | 0.25->64 | 8 | 32 | ||
| Tetracycline | 8->64 | >64 | >64 | ||
| All Tet-resistant isolates | 488 | Tigecycline | 0.06-2 | 0.25 | 0.5 |
| Minocycline | 0.25->64 | 8 | 32 | ||
| Tetracycline | 8->64 | >64 | >64 | ||
| Tetracycline-susceptible isolates | 727 | Tigecycline | 0.03-1 | 0.25 | 0.5 |
| Minocycline | 0.12-4 | 1 | 2 | ||
| Tetracycline | 0.12-4 | 2 | 4 | ||
| E. coli ATCC 25922 | Tigecycline | 0.125-5 | NA | NA | |
| Minocycline | 0.5-1 | NA | NA | ||
| Tetracycline | ND | ND | ND | ||
NA, not applicable—less than 10 strains analyzed; ND, not determined.
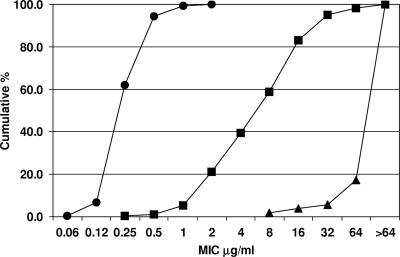
FIG. 1.
Susceptibility profile of 488 tetracycline-resistant E. coli isolates. The figure shows the cumulative percentage of strains versus the respective MICs of tetracycline (▴), minocycline (▪), and tigecycline (•). The leftward shift for the minocycline trace is indicative of the subset of isolates that fail to efflux minocycline; the further leftward shift of the tigecycline trace underlies the inability of classical tetracycline resistance determinants to circumvent tigecycline.
All isolates were susceptible to tigecycline, with an MIC range of 0.06 to 2 μg/ml and an MIC90 of 0.5 μg/ml, regardless of the type or number of expressed efflux pumps encoded or the presence of the tet(M) determinant (Table 3; Fig. 1). The MIC50 and MIC90 were 0.25 μg/ml and 0.5 μg/ml, respectively, for single-determinant as well as multiple-determinant-encoding isolates.
DISCUSSION
Tetracycline-specific efflux pump proteins, members of the MFS family of pumps, are the leading tetracycline resistance mechanisms in E. coli (10, 28, 33). In prior clinical surveys, the tet(B) gene was the most prevalent tetracycline resistance determinant identified, having a wide host range due to the fact that it resides on highly mobile genetic elements that readily transfer between different bacterial genera (31, 41). The tet(A), tet(C), and tet(D) genes are located on conjugative plasmids of different incompatibility groups (20), whereas the tet(E) determinant has been located on the chromosome in some isolates and has also associated with large, nonconjugative, nonmobile plasmids (30).
E. coli strains collected during the tigecycline clinical trials showed an incidence of tetracycline resistance similar (39%) to those in recently reported worldwide surveillance studies (30 to 45%) (16, 17, 35, 36). In the current study, a large percentage of the resistant strains (33%) encoded multiple tetracycline efflux pumps. This confirms the recent upward trend in which >30% of environmental isolates were found to encode multiple tetracycline efflux pumps (6, 40), an increase of more than three times that reported recently from other sources (33). A correlation between the number of encoded tetracycline resistance genes and the level of tetracycline resistance (in MICs) has not been noted. In addition to the increasing burden of tetracycline in the environment, the increased use of various biocides, i.e., triclosan and chlorhexidine, may be responsible for the increase in the frequency of isolates encoding multiple tetracycline resistance determinants (4, 23). The continued genetic exchange of resistance determinants among various environmental, commensal, and clinical bacteria would also be expected to have a clinical impact.
There were 16 isolates in the strain collection that, although PCR positive for tet(B), were susceptible to minocycline (MIC, ≤4 μg/ml) and resistant to tetracycline (MIC, ≥64 μg/ml). Of these isolates, 13 encoded an additional tet determinant, tet(A) or tet(E). Therefore, it is possible that the tet(B) gene is either poorly expressed or not expressed in these strains. For the three strains that expressed only tet(B) and resulted in an MIC of ≤4 μg/ml for minocycline and an MIC of >64 μg/ml for tetracycline, it may be that the level of expression is too low to result in overt resistance to minocycline or that tetracycline resistance in these isolates is mediated by a gene not included in our PCR panel.
Somewhat surprisingly, 36 isolates in this clinical collection were negative for tet(A) to tet(E) and tet(M), as well as tet(G), tet(K), tet(L), and tet(Y). One possible explanation is that a number of these isolates encode a mosaic gene encoding a ribosomal protection protein, as has recently been described in a number of human and animal fecal samples (27). Our primer sets were not designed to capture these unique genetic elements that therefore would have been missed in this analysis.
Four E. coli isolates were collected that tested with a tigecycline MIC of 2 μg/ml. Two of these isolates were negative for all tet determinants on the PCR screening panel, one isolate encoded tet(A), and the fourth isolate was shown to encode tet(A) plus tet(D). As 24 other isolates encoded this combination and this group of strains had an MIC90 of 1 μg/ml, there was no correlation between this combination of efflux pumps and reduced susceptibility to tigecycline. Recent studies have implicated the overexpression of AcrAB, a member of the RND multidrug efflux family, in E. coli as contributing to reduced susceptibility to tigecycline (D. Keeney, A. Ruzin, F. McAleese, and P. A. Bradford, submitted for publication).
Tigecycline, a new broad-spectrum antibiotic, is the first antibiotic of the glycylcycline class approved for clinical use (Tygacil package insert, Wyeth Pharmaceuticals Inc., Collegeville, PA). Tigecycline evades both the ribosomal protection and efflux pump resistance mechanisms that are the two most common mechanisms of tetracycline resistance in clinical strains (15, 29). In vitro studies have demonstrated that tigecycline is not a substrate for the TetB efflux pump and does not induce proton transport across membranes (19). Ribosomal binding studies have demonstrated that tigecycline binds more tightly than tetracycline and minocycline to the ribosome; therefore, tigecycline may be less efficiently displaced from the 30S ribosomal subunit by ribosomal protection proteins than the classical tetracyclines (3, 26). The results of this study confirm that the presence of one or more tetracycline resistance determinants does not affect tigecycline susceptibility in clinical isolates of E. coli.
These studies provide a current (2002 to 2006) picture of the burden of tetracycline resistance determinants among clinical E. coli isolates, as well as the utility of the novel broad-spectrum agent, tigecycline, against these pathogens. Moreover, these results support the general approach of reengineering existing antimicrobial agents with acceptable safety profiles to evade the resistance mechanisms posed by bacterial pathogens.
Footnotes
Published ahead of print on 9 July 2007.
REFERENCES
- 1.Akiba, T., K. Koyama, Y. Ishiki, S. Kimura, and T. Fukushima. 1960. On the mechanism of the development of multiple-drug-resistant clones of Shigella. Jpn. J. Microbiol. 4:219-227. [DOI] [PubMed] [Google Scholar]
- 2.Babinchak, T., E. J. Ellis-Grosse, N. Dartois, G. M. Rose, and E. G. Loh. 2005. The efficacy and safety of tigecycline in the treatment of complicated intra-abdominal infections: analysis of pooled clinical trial data. Clin. Infect. Dis. 41:S354-S367. [DOI] [PubMed] [Google Scholar]
- 3.Bauer, G., C. Berens, S. J. Projan, and W. Hillen. 2004. Comparison of tetracycline and tigecycline binding to ribosomes mapped by dimethylsulphate and drug-directed Fe2+ cleavage of 16S rRNA. J. Antimicrob. Chemother. 53:592-599. [DOI] [PubMed] [Google Scholar]
- 4.Braoudaki, M., and A. C. Hilton. 2004. Adaptive resistance to biocides in Salmonella enterica and Escherichia coli O157 and cross-resistance to antimicrobial agents. J. Clin. Microbiol. 42:73-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Briggs, C. E., and P. M. Fratamico. 1999. Molecular characterization of an antibiotic resistance gene cluster in Salmonella typhimurium DT 104. Antimicrob. Agents Chemother. 43:846-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bryan, A., N. Shapir, and M. J. Sadowsky. 2004. Frequency and distribution of tetracycline resistance genes in genetically diverse, nonselected, and nonclinical Escherichia coli strains isolated from diverse human and animal sources. Appl. Environ. Microbiol. 70:2503-2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burdett, V. 1986. Streptococcal tetracycline resistance mediated at the level of protein synthesis. J. Bacteriol. 165:564-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Charpentier, E., G. Gerbaud, and P. Courvalin. 1994. Presence of the Listeria tetracycline resistance gene tet(S) in Enterococcus faecalis. Antimicrob. Agents Chemother. 38:2330-2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chopra, I. 2002. New developments in tetracycline antibiotics: glycylcyclines and tetracycline efflux pump inhibitors. Drug Resist. Update 5:119-125. [DOI] [PubMed] [Google Scholar]
- 10.Chopra, I., and M. Roberts. 2001. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 65:232-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clewell, D. B., S. E. Flannagan, and D. D. Jaworski. 1995. Unconstrained bacterial promiscuity: the Tn916-Tn1545 family of conjugative transposons. Trends Microbiol. 3:229-236. [DOI] [PubMed] [Google Scholar]
- 12.CLSI. 2006. Performance standards for antimicrobial susceptibility testing; 16th informational supplement, vol. 26. M100-S16. Clinical and Laboratory Standards Institute, Wayne, PA.
- 13.Eisenstein, B. I., and D. F. Zaleznik. 2000. Enterobacteriaceae, p. 2294-2310. In G. L. Mandell, J. E. Bennett, and R. Dolin (ed.), Principles and practice of infectious diseases, 5th ed., vol. 2. Churchill Livingstone, Philadelphia, PA. [Google Scholar]
- 14.Ellis-Grosse, E., T. Babinchak, N. Dartois, G. Rose, and E. Loh. 2005. The efficacy and safety of tigecycline in the treatment of skin and skin structure infections: results of two double-blind phase 3 comparison studies with vancomycin/aztreonam. Clin. Infect. Dis. 41:S341-S353. [DOI] [PubMed] [Google Scholar]
- 15.Fluit, A. C., A. Florijn, J. Verhoef, and D. Milatovic. 2005. Presence of tetracycline resistance determinants and susceptibility to tigecycline and minocycline. Antimicrob. Agents Chemother. 49:1636-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fritsche, T. R., H. S. Sader, M. G. Stilwell, M. J. Dowzicky, and R. N. Jones. 2005. Potency and spectrum of tigecycline tested against an international collection of bacterial pathogens associated with skin and soft tissue infections (2000-2004). Diagn. Microbiol. Infect. Dis. 52:195-201. [DOI] [PubMed] [Google Scholar]
- 17.Fritsche, T. R., P. A. Strabala, H. S. Sader, M. J. Dowzicky, and R. N. Jones. 2005. Activity of tigecycline tested against a global collection of Enterobacteriaceae, including tetracycline-resistant isolates. Diagn. Microbiol. Infect. Dis. 52:209-213. [DOI] [PubMed] [Google Scholar]
- 18.Guay, G. G., and D. M. Rothstein. 1993. Expression of the tetK gene from Staphylococcus aureus in Escherichia coli: comparison of substrate specificities of TetA(B), TetA(C), and TetK efflux proteins. Antimicrob. Agents Chemother. 37:191-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hirata, T., A. Saito, K. Nishino, N. Tamura, and A. Yamaguchi. 2004. Effects of efflux transporter genes on susceptibility of Escherichia coli to tigecycline (GAR-936). Antimicrob. Agents Chemother. 48:2179-2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones, C. E. S., D. J. Osborne, and J. Stanley. 1992. Enterobacterial tetracycline resistance in relation to plasmid incompatibility. Mol. Cell. Probes 6:313-317. [DOI] [PubMed] [Google Scholar]
- 21.Jones, C. H., M. Tuckman, A. Y. M. Howe, M. Orlowski, S. Mullen, K. Chan, and P. A. Bradford. 2006. Diagnostic PCR analysis of the occurrence of methicillin and tetracycline resistance genes among Staphylococcus aureus isolates from phase 3 clinical trials of tigecycline for complicated skin and skin structure infections. Antimicrob. Agents Chemother. 50:505-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones, C. H., M. Tuckman, E. Murphy, and P. A. Bradford. 2006. Identification and sequence of a tet(M) tetracycline resistance determinant homologue in clinical isolates of Escherichia coli. J. Bacteriol. 188:7151-7164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levy, S. B. 2002. Active efflux, a common mechanism for biocide and antibiotic resistance. Symp. Ser. Soc. Appl. Microbiol. 31:65S-71S. [PubMed] [Google Scholar]
- 24.Levy, S. B. 2001. Antibiotic resistance: consequences of inaction. Clin. Infect. Dis. 33(Suppl. 3):S124-S129. [DOI] [PubMed] [Google Scholar]
- 25.Ojo, K. K., N. L. Ruehlen, N. S. Close, H. Luis, M. Bernardo, J. Leitao, and M. C. Roberts. 2006. The presence of a conjugative gram-positive Tn2009 in gram-negative commensal bacteria. J. Antimicrob. Chemother. 57:1065-1069. [DOI] [PubMed] [Google Scholar]
- 26.Olson, M. W., A. Ruzin, E. Feyfant, T. S. Rush III, J. F. O'Connell, and P. A. Bradford. 2006. Functional, biophysical, and structural bases for antibacterial activity of tigecycline. Antimicrob. Agents Chemother. 50:2156-2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patterson, A. J., M. T. Rincon, H. J. Flint, and K. P. Scott. 2007. Mosaic tetracycline resistance genes are widespread in human and animal fecal samples. Antimicrob. Agents Chemother. 51:1115-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paulsen, I. T., M. H. Brown, and R. A. Skurray. 1996. Proton-dependent multidrug efflux systems. Microbiol. Rev. 60:575-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Petersen, P. J., N. V. Jacobus, W. J. Weiss, P. E. Sum, and R. T. Testa. 1999. In vitro and in vivo antimicrobial activities of a novel glycylcycline, the 9-t-butylglycylamido derivative of minocycline (GAR-936). Antimicrob. Agents Chemother. 43:738-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roberts, M. C. 1994. Epidemiology of tetracycline-resistance determinants. Trends Microbiol. 2:353-357. [DOI] [PubMed] [Google Scholar]
- 31.Roberts, M. C. 1996. Tetracycline resistance determinants: mechanisms of action, regulation of expression, genetic mobility, and distribution. FEMS Microbiol. Rev. 19:1-24. [DOI] [PubMed] [Google Scholar]
- 32.Roberts, M. C. 2003. Tetracycline therapy: update. Clin. Infect. Dis. 36:462-467. [DOI] [PubMed] [Google Scholar]
- 33.Roberts, M. C. 2005. Update on acquired tetracycline resistance genes. FEMS Microbiol. Lett. 245:195-203. [DOI] [PubMed] [Google Scholar]
- 34.Ross, J. I., E. A. Eady, J. H. Cove, and W. J. Cunliffe. 1998. 16S rRNA mutation associated with tetracycline resistance in a gram-positive bacterium. Antimicrob. Agents Chemother. 42:1702-1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sader, H. S., R. N. Jones, M. J. Dowzicky, and T. R. Fritsche. 2005. Antimicrobial activity of tigecycline tested against nosocomial bacterial pathogens from patients hospitalized in the intensive care unit. Diagn. Microbiol. Infect. Dis. 52:203-208. [DOI] [PubMed] [Google Scholar]
- 36.Sader, H. S., R. N. Jones, M. G. Stilwell, M. J. Dowzicky, and T. R. Fritsche. 2005. Tigecycline activity tested against 26,474 bloodstream infection isolates: a collection from 6 continents. Diagn. Microbiol. Infect. Dis. 52:181-186. [DOI] [PubMed] [Google Scholar]
- 37.Schnabel, E. L., and A. L. Jones. 1999. Distribution of tetracycline resistance genes and transposons among phylloplane bacteria in Michigan apple orchards. Appl. Environ. Microbiol. 65:4898-4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smalla, K., H. Heuer, A. Gotz, D. Niemeyer, E. Krogerrecklenfort, and E. Tietze. 2000. Exogenous isolation of antibiotic resistance plasmids from piggery manure slurries reveals a high prevalence and diversity of IncQ-like plasmids. Appl. Environ. Microbiol. 66:4854-4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tietze, E. 1998. Nucleotide sequence and genetic characterization of the novel IncQ plasmid pIE 1107. Plasmid 39:165-181. [DOI] [PubMed] [Google Scholar]
- 40.Villedieu, A., M. L. Diaz-Torres, N. Hunt, R. McNab, D. A. Spratt, M. Wilson, and P. Mullany. 2003. Prevalence of tetracycline resistance genes in oral bacteria. Antimicrob. Agents Chemother. 47:878-882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wilkerson, C., M. Samadpour, N. van Kirk, and M. C. Roberts. 2004. Antibiotic resistance and distribution of tetracycline resistance genes in Escherichia coli O157:H7 isolates from humans and bovines. Antimicrob. Agents Chemother. 48:1066-1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang, W., I. F. Moore, K. P. Koteva, D. C. Bareich, D. W. Hughes, and G. D. Wright. 2004. TetX is a flavin-dependent monooxygenase conferring resistance to tetracycline antibiotics. J. Biol. Chem. 279:52346-52352. [DOI] [PubMed] [Google Scholar]