Abstract
Eukaryotic cells have evolved molecular mechanisms to ensure the faithful inheritance of organelles by daughter cells in order to maintain the benefits afforded by the compartmentalization of biochemical functions. Little is known about the inheritance of peroxisomes, organelles of lipid metabolism. We have analyzed peroxisome dynamics and inheritance in the dimorphic yeast Yarrowia lipolytica. Most peroxisomes are anchored at the periphery of cells of Y. lipolytica. In vivo video microscopy showed that at cell division, approximately half of the anchored peroxisomes in the mother cell are dislodged individually from their static positions and transported to the bud. Peroxisome motility is dependent on the actin cytoskeleton. YlInp1p is a peripheral peroxisomal membrane protein that affects the partitioning of peroxisomes between mother cell and bud in Y. lipolytica. In cells lacking YlInp1p, most peroxisomes were transferred to the bud, with only a few remaining in the mother cell, while in cells overexpressing YlInp1p, peroxisomes were preferentially retained in the mother cell, resulting in buds nearly devoid of peroxisomes. Our results are consistent with a role for YlInp1p in anchoring peroxisomes in cells. YlInp1p has a role in the dimorphic transition in Y. lipolytica, as cells lacking the YlINP1 gene more readily convert from the yeast to the mycelial form in oleic acid-containing medium, the metabolism of which requires peroxisomal activity, than does the wild-type strain. This study reports the first analysis of organelle inheritance in a true dimorphic yeast and identifies the first protein required for peroxisome inheritance in Y. lipolytica.
Eukaryotic cells have evolved highly regulated and coordinated molecular mechanisms to ensure the accurate partitioning of their organelle populations between mother and daughter cells at cell division. These mechanisms involve molecular motors that transport organelles between mother and daughter cells along cytoskeletal elements, specific organellar receptors that recognize the molecular motors, and proteins that retain organelles within both mother cell and daughter to ensure an equitable division of organelles between them (16, 31-33, 35).
Peroxisomes are ubiquitous organelles involved in a myriad of biochemical processes, most notably the oxidative metabolism of fatty acids and the decomposition of hydrogen peroxide (2, 8, 11, 15, 29). Peroxisomes arise by both de novo biogenesis at the endoplasmic reticulum and growth and division of preexisting peroxisomes (for recent reviews, see references 10, 14, 22, 25, and 26). Peroxisomes are also inherited at cell division (4). During cell division of the budding yeast Saccharomyces cerevisiae, approximately half of the peroxisome population of a mother cell is transferred to the bud along actin cables by the class V myosin motor Myo2p, while the other half is retained at the mother cell cortex (3, 5, 7). The peroxisomal membrane protein Inp1p (inheritance of peroxisomes protein 1) was the first bona fide peroxisomal protein shown to be required for peroxisome inheritance (5). Inp1p acts to retain peroxisomes in both the mother cell and bud and has been proposed to link peroxisomes to an as-yet-unidentified anchoring structure at the cell cortex (5). Recently, the peroxisomal membrane protein Inp2p was shown to be the peroxisome-specific receptor for Myo2p (3).
Yarrowia lipolytica is a dimorphic, heterothallic, ascomycetous yeast that has been used industrially for many years for the exogenous production of protein (13). Y. lipolytica has also proven to be an excellent model system with which to identify and characterize the processes and molecules of the peroxisome biogenic program, particularly in regards to the mechanisms of recognition and import of proteins destined for the peroxisome and of de novo synthesis of peroxisomes from the endoplasmic reticulum (for a review, see reference 30). However, it is unclear whether peroxisomes can also be inherited in Y. lipolytica and what might the mechanisms and proteins be that would underlie this process. Here we report that peroxisome movement and inheritance in Y. lipolytica are dependent on actin and that Inp1p of S. cerevisiae shares sequence homology with a protein encoded by the open reading frame YALI0F31229g of the Y. lipolytica genome. YALI0F31229g codes for a peroxisomal peripheral membrane protein, which we term YlInp1p, that is required for peroxisome inheritance in Y. lipolytica and is also involved in the dimorphic transition in this yeast.
MATERIALS AND METHODS
Strains and culture conditions.
The Y. lipolytica strains used in this study are listed in Table 1. Strains were cultured at 30°C. Strains containing plasmids were cultured in minimal medium (YND or YNO). All other strains were cultured in YPD or YPBO medium. Strains expressing genomically encoded green fluorescent protein (GFP) chimeras were constructed by homologous recombination using a PCR-based integrative transformation. Media components were as follows: YPD, 1% yeast extract, 2% peptone, 2% glucose; YPBO, 0.3% yeast extract, 0.5% peptone, 0.5% K2HPO4, 0.5% KH2PO4, 1% Brij 35, 1% oleic acid; YND, 0.67% yeast nitrogen base without amino acids, 2% glucose; YNO, 0.67% yeast nitrogen base without amino acids, 0.05% Tween 40, 0.1% oleic acid. YND and YNO were supplemented with leucine, lysine, and uracil, each at 50 μg/ml, as required.
TABLE 1.
Y. lipolytica strains used in this study
| Strain | Genotype |
|---|---|
| E122a | MATaura3-302 leu2-270 lys8-11 |
| Ylinp1Δ | MATaura3-302 leu2-270 lys8-11 Ylinp1::URA3 |
| E122/POT1-GFP | MATaura3-302 leu2-270 lys8-11 pot1::POT1-GFP(LEU2) |
| Ylinp1Δ/POT1-GFP | MATaura3-302 leu2-270 lys8-11 Ylinp1::URA3 pot1::POT1-GFP(LEU2) |
| YlINP1-GFP | MATaura3-302 leu2-270 lys8-11 Ylinp1::YlINP1-GFP(LEU2) |
| TC3-YlINP1/ | |
| POT1-GFP | MATaura3-302 leu2-270 lys8-11 pTC3(URA3)YlINP1 pot1::POT1-GFP(LEU2) |
A gift of C. Gaillardin (Thiverval-Grignon, France). All other strains were from this study.
Integrative disruption of the YlINP1 gene.
Targeted integrative disruption of the YlINP1 gene of Y. lipolytica was performed by overlapping PCR (1). A 1.7-kbp fragment containing the Y. lipolytica URA3 gene and 250 bp at both the 5′ end and 3′ end of the open reading frame of the YlINP1 gene were amplified by PCR. The three amplified products were then used as templates for overlapping PCR to produce a fragment containing the YlURA3 gene flanked at its 5′ and 3′ ends by ∼250 bp of the YlINP1 open reading frame. This fragment was used to transform Y. lipolytica wild-type strain E122 to uracil prototrophy. Ura+ transformants were selected and screened by PCR amplification for correct integration of the YlURA3 gene into the YlINP1 locus.
Staining cell structures.
Actin was stained with rhodamine-phalloidin (Molecular Probes). Vacuoles were stained with N-(3-triethylammoniumpropyl)-(6(4 (diethylamino)phenyl)hexatrienyl) pyridium dibromide (FM4-64; Molecular Probes). Mitochondria were stained with MitoTracker Red CMXRos (Molecular Probes).
Plasmids.
The Y. lipolytica expression plasmid pTC3 containing a unique EcoRI site between the promoter and terminator regions of the POT1 gene encoding the peroxisomal matrix enzyme 3-ketoacyl-coenzyme A (CoA) thiolase (Pot1p) has been described elsewhere (12). The plasmid pTC3-mRFP-SKL encoding monomeric red fluorescent protein (mRFP) tagged at its carboxyl terminus with the peroxisome targeting signal 1 (PTS1), Ser-Lys-Leu, was a kind gift of Yuen Yi Chris Tam (University of Alberta). To overexpress YlINP1, its open reading frame was amplified by PCR and inserted into pTC3 to make pTC3-YlINP1.
4D in vivo video microscopy.
Four-dimensional (4D) in vivo video microscopy was performed essentially as described previously (5). Cells synthesizing a genomically encoded chimera between Pot1p and GFP (Pot1p-GFP) were grown in YPD medium and then incubated in YPBO medium for 16 h. Cells were placed in a chambered no. 1.0 borosilicate coverglass (Lab Tek) coated with concanavalin A and incubated at room temperature for image capture. Images were captured with a Plan-Apochromat 63×, 1.4 numerical-aperture oil differential interference contrast objective and an Axiovert 200 microscope equipped with an LSM 510 META confocal scanner (Carl Zeiss). A piezoelectric actuator was used to drive continuous objective movement, allowing for the rapid collection of z-stacks. The sides of each pixel represent 0.085 μm of the sample. Stacks of 13 (see video S4 in the supplemental material), 14 (see videos S5, S6, and S7 in the supplemental material), 15 (see video S2), or 16 (see videos S1 and S3) optical sections spaced 0.45 μm apart were captured every 12 s or every 2 min (video S4 only). GFP was excited using a 488-nm laser, and its emission was collected using a 505-nm long-pass filter. The resulting images were filtered three times using a 3 by 3 hybrid median filter to reduce shot noise. Fluorescent images from each stack were projected using an average intensity algorithm that involved multiplication of each pixel value by an appropriate enhancement factor for better contrast. Correction for exponential photobleaching of GFP was performed by exponentially increasing the enhancement factor with each projection. The transmitted light images from each stack were projected using a maximum intensity algorithm. The resulting projections were smoothed by means of a blurring algorithm. These operations were performed using NIH Image (http://rsb.info.nih.gov/nih-image/). Adobe Photoshop (Adobe Systems) was used to merge the fluorescent and transmitted light projections. Processed projections were assembled into videos using Apple QuickTime Pro 6.5.2 at a rate of either 10 frames per second or 6 frames per second (video S4 only). Postprocessing operations such as peroxisome tracking and 3D reconstruction were performed using Imaris 4.1 (Bitplane).
Quantification of peroxisome inheritance.
Peroxisome inheritance was quantified essentially as previously described for S. cerevisiae (5). Cells were grown in YPD medium for 16 h and then transferred to YPBO medium for 16 h. Peroxisomes were visualized by direct fluorescence confocal microscopy. For each randomly chosen field, three optical sections of 5-μm thickness were collected at a z-axis spacing of 1.6 μm using a high detector gain to ensure the capture of weak fluorescent signals. Quantification of peroxisome inheritance in wild-type and Ylinp1Δ cells was done by determining the percentage of mother cells retaining peroxisomes at their tips distal to the site of bud growth and in cells overexpressing YlINP1 by determining the percentage of buds that retain peroxisomes at their tips and that are at least half the size of their corresponding mother cell. Quantification was performed on 100 budded cells of each strain.
Cell fractionation and peroxisome subfractionation.
Fractionation of oleic acid-induced cells was performed essentially as described previously (21). Homogenized spheroplasts were subjected to differential centrifugation at 1,000 × g for 10 min at 4°C in a JS13.1 rotor (Beckman) to yield a postnuclear supernatant fraction. The postnuclear supernatant fraction was subjected to further differential centrifugation at 20,000 × g for 30 min at 4°C to yield a pellet (20KgP) fraction enriched for peroxisomes and mitochondria and a supernatant (20KgS) fraction enriched for cytosol. Peroxisomes were purified from the 20KgP fraction by isopycnic centrifugation on a discontinuous sucrose gradient (27). Peroxisomes were separated into fractions enriched for matrix, peripheral, and integral membrane proteins by treatment with dilute Tris and alkali Na2CO3, as described elsewhere (20).
Antibodies.
Antibodies to YlInp1p were raised in guinea pigs against a glutathione S-transferase/YlInp1p fusion. The entire open reading frame of YlINP1 was amplified by PCR and cloned into the plasmid pGEX-4T-1 (Amersham Biotech) downstream and in frame to the open reading frame for glutathione S-transferase, followed by expression in Escherichia coli. Anti-YlInp1p serum was partially purified by immunodepletion versus a lysate of Ylinp1Δ cells. Antibodies to Pex2p, Pot1p, and Sdh2p have been described previously (23, 24). Antitubulin antibodies raised in rat and rhodamine-conjugated guinea pig anti-rat immunoglobulin G (IgG) secondary antibodies were kind gifts of Neil Adames (University of Alberta). Horseradish peroxidase-conjugated donkey anti-rabbit IgG and horseradish peroxidase-conjugated goat anti-guinea pig IgG secondary antibodies were used to detect primary antibodies in immunoblot analysis by enhanced chemiluminescence (Amersham Bioscience).
RESULTS
Peroxisome dynamics in wild-type Y. lipolytica cells.
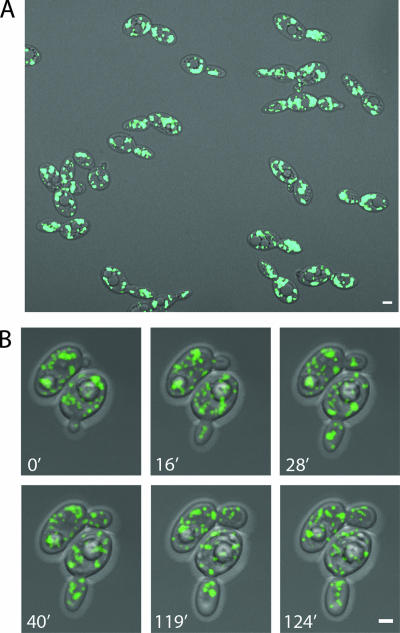
To begin to investigate peroxisome inheritance in Y. lipolytica, we first observed the distribution and movement of peroxisomes in wild-type E122 cells by using 4D in vivo video microscopy. The peroxisomal β-oxidation enzyme 3-ketoacyl-CoA thiolase (Pot1p) was genomically tagged with GFP (Pot1p-GFP) by homologous recombination with a PCR-based integrative transformation of E122 to make the strain E122/POT1-GFP, in which peroxisomes are fluorescently labeled. Incubation of E122/POT1-GFP cells for 16 h in YPBO medium containing oleic acid, the metabolism of which requires functional peroxisomes, yielded 30 to 40 fluorescent punctate structures characteristic of peroxisomes per cell on average (Fig. 1A). Peroxisomes in both mother cells and buds were observed by video microscopy to be highly mobile and to exhibit both anterograde and retrograde movement (Fig. 1B; see also video S1 in the supplemental material). The maximum velocities of anterograde and retrograde peroxisome movement were determined to be approximately the same (0.41 μm/s and 0.39 μm/s, respectively). Peroxisomes in the mother cell were seen to oscillate at the cell cortex for a period and then suddenly alter their positions. Peroxisomes would often gather and then separate. When peroxisomes entered the bud from the mother cell, they would first arrive at the middle of the bud and, following a small amount of movement there, move to the bud tip and then return to the middle of the bud. Peroxisomes were also frequently observed to return from the bud to the mother cell, an event never observed in wild-type S. cerevisiae cells (3, 5).
FIG. 1.
Peroxisome dynamics in Y. lipolytica. (A) Wild-type cells expressing genomically encoded Pot1p-GFP to fluorescently label peroxisomes were grown in glucose-containing YPD medium and then transferred to oleic acid-containing YPBO medium and incubated for 16 h. Fluorescent images were captured by confocal microscopy. In this static picture, peroxisomes are seen to be evenly distributed between mother cell and bud. (B) The strain used in panel A was analyzed by 4D in vivo video microscopy. Representative frames from video S1 in the supplemental material show the specific movements of peroxisomes. One peroxisome already present in the small bud of the cell at bottom at the start of the video (0′) was observed later to return to the mother cell. At 16 min, one peroxisome in the cell at the top entered the bud. Additional peroxisomes entered this bud one by one between 16 min and 90 min. During this period, peroxisomes in the bud sometimes clustered at the center of the bud (28′) and later separated (40′), and some peroxisomes returned to the mother cell. Before cytokinesis, some peroxisomes from both the mother cell and bud relocated to the mother-bud neck region (119′) and then redistributed (124′). Bars, 2 μm.
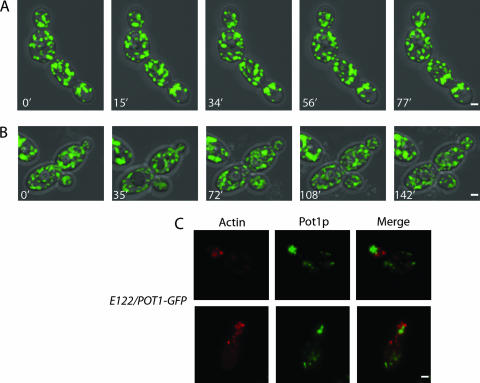
Peroxisome movement in Y. lipolytica is dramatically reduced by depolymerization of actin but not microtubules.
Peroxisome movement involves actin in the yeast S. cerevisiae and in plants but uses microtubules in mammalian cells. When E122/POT1-GFP cells were treated with the actin-disrupting toxin latrunculin A, peroxisomes exhibited greatly reduced movement and oscillated in position at the cortex of both mother cell and bud (Fig. 2A; see also video S2 in the supplemental material). In contrast, treatment of cells with the microtubule inhibitor nocodazole seemingly did not affect peroxisome movement, although by 120 min of treatment, cells usually arrested with a single large bud (Fig. 2B and unpublished data; see also video S3 in the supplemental material). Staining of cells with rhodamine-phalloidin to detect actin and with antitubulin antibodies confirmed disruption of actin and microtubules by latrunculin A and nocodazole, respectively (unpublished results).
FIG. 2.
Actin is involved in peroxisome dynamics. Treatment of cells with the actin-disrupting toxin latrunculin A (A) but not with the microtubule inhibitor nocodazole (B) abolished the dynamic movements of peroxisomes. Although cells grew more slowly following treatment with nocodazole, peroxisomes were still recruited to buds as normal. Representative frames from videos S2 and S3 in the supplemental material are presented in panels A and B, respectively. (C) Peroxisomes do not colocalize with actin patches. Wild-type cells synthesizing genomically encoded Pot1p-GFP were grown in YPD medium and then transferred to YPBO medium for 16 h. Actin was detected by staining with rhodamine-phalloidin and visualized by epifluorescence microscopy. Bar, 2 μm.
In S. cerevisiae, the actin cytoskeleton consists of distinct patches and cables that orient toward the bud neck and tip. In filamentous fungi such as Aspergillus nidulans and Neurospora crassa, actin cables are not observed; however, actin patches that localize to actively growing or emerging hyphal tips and at sites of septation are seen (34). In Y. lipolytica, actin cables are also not observed, but actin patches are present, and their distribution during the cell cycle is similar to that described for actin patches in S. cerevisiae (6; unpublished data). E122/POT1-GFP cells stained with rhodamine-phalloidin showed actin patches at sites of polarized growth (Fig. 2C). Peroxisomes were closely apposed to, but did not colocalize with, actin patches (Fig. 2C).
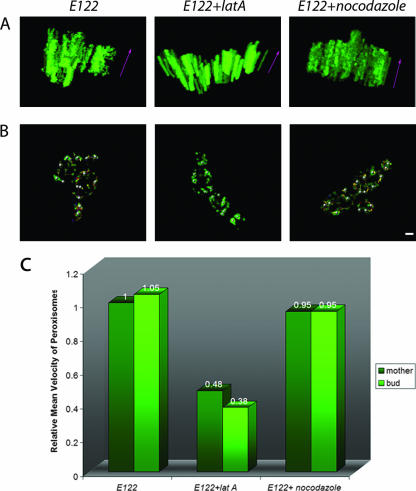
The movement of peroxisomes in wild-type, latrunculin A-treated, and nocodazole-treated cells was also analyzed using 3D kymographs (Fig. 3A), which were constructed by overlapping the last 100 projections, corresponding to 20 min of real time in videos S1, S2, and S3 in the supplemental material, respectively. Static peroxisomes are seen as fluorescent columns, while mobile peroxisomes are seen as fluorescent spots that change position with time. Mobile peroxisomes were often observed in wild-type and nocodazole-treated cells, while latrunculin A-treated cells showed mostly fluorescent columns, each indicative of a static peroxisome (Fig. 3A). The tracking of randomly selected, individual peroxisomes in wild-type, latrunculin A-treated, and nocodazole-treated cells during the last 20 min of videos S1, S2, and S3 is presented in Fig. 3B.
FIG. 3.
Quantification of peroxisome mobility. (A) One hundred projections corresponding to the last 20 min of videos S1, S2, and S3 in the supplemental material were analyzed with Imaris 4.1 (Bitplane), and 3D models were constructed. The z axis (purple arrows) represents time. A peroxisome that maintains its x-y position for the period of time considered and which is essentially immobile is represented by a fluorescent column parallel to the z axis. A mobile peroxisome is represented by fluorescent spots that have different x-y positions in time. (B) Tracking peroxisomes in untreated, latrunculin A-treated, and nocodazole-treated wild-type E122 cells. Randomly selected peroxisomes under each condition were tracked by analyzing the last 100 projections of videos S1, S2, and S3 with Imaris 4.1. The trajectories of individual peroxisomes are shown as different-colored lines. Bar, 2 μm. (C) Peroxisomes in latrunculin A-treated cells exhibit reduced mobility. The velocities of individual peroxisomes across individual time points were measured using Imaris 4.1, and an average velocity was obtained for each peroxisome. The average velocities of individual peroxisomes under a given condition were in turn averaged to obtain the mean velocity of peroxisomes under that condition. The mean velocity of peroxisomes under a given condition is expressed relative to the mean velocity of peroxisomes of the untreated wild-type E122/POT1-GFP strain, which was taken as 1.
We calculated the mean velocities of peroxisomes in wild-type, latrunculin A-treated, and nocodazole-treated cells (Fig. 3C). The mean velocities of peroxisomes in wild-type and nocodazole-treated cells were similar and were approximately twice that of peroxisomes in latrunculin A-treated cells. Within each group of cells, there was no difference in mean velocity between mother cell and bud (Fig. 3C).
YlInp1p is a peripheral peroxisomal protein.
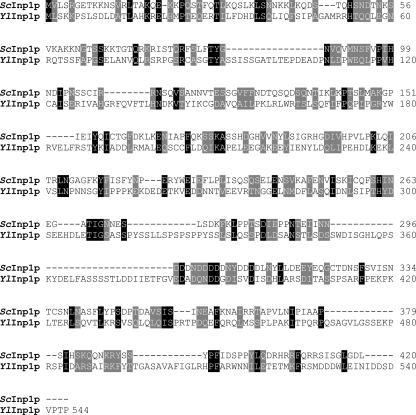
A requirement for actin in peroxisomal movement within and between cells of Y. lipolytica led us to speculate that Y. lipolytica might use mechanisms for peroxisome inheritance similar to those used by S. cerevisiae. Inp1p is a peripheral membrane protein of peroxisomes that was recently shown to be required for peroxisome inheritance in S. cerevisiae (5). A search of protein databases with the GENEINFO BLAST network service of the National Center for Biotechnology Information revealed one protein encoded by open reading frame YALI0F31229g of the Y. lipolytica genome with sequence similarity to Inp1p. Hereafter, the protein encoded by open reading frame YALI0F31229g is termed YlInp1p and its encoding gene as YlINP1. Inp1p and YlInp1p exhibit 11.6% amino acid identity and 19.9% amino acid similarity (Fig. 4).
FIG. 4.
Sequence alignment of S. cerevisiae Inp1p with the hypothetical YlInp1p encoded by the open reading frame YALI0F31229g of the Y. lipolytica genome. Amino acid sequences were aligned with the use of the ClustalW program (http://www.ebi.ac.uk/clustalw/; EMBL-EBI, Cambridgeshire, United Kingdom). Identical residues (black) and similar residues (gray) in the two proteins are shaded. Similarity rules: G = A = S; A = V; V = I = L = M; I = L = M = F = Y = W; K = R = H; D = E = Q = N; S = T = Q = N. Dashes represent gaps.
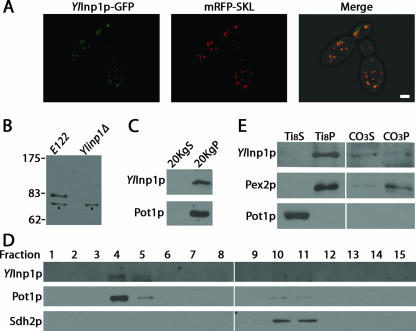
Confocal fluorescence microscopy was used to determine the localization of a genomically encoded fluorescent chimera of YlInp1p and GFP (YlInp1p-GFP). Peroxisomes were decorated by a plasmid-encoded fluorescent chimera (mRFP-SKL) of mRFP and the peroxisome targeting signal 1, Ser-Lys-Leu (SKL). YlInp1p-GFP colocalized with mRFP-SKL to punctate structures characteristic of peroxisomes (Fig. 5A).
FIG. 5.
YlInp1p is a peripheral membrane protein of peroxisomes. (A) YlInp1p-GFP colocalizes with mRFP-SKL to punctate structures characteristic of peroxisomes by direct confocal microscopy. The right panel presents the merged image of the left and middle panels, with colocalization of YlInp1p-GFP and mRFP-SKL shown in yellow. Bar, 2 μm. (B) Immunoblot analysis of whole-cell lysates of the wild-type strain E122 and the deletion strain Ylinp1Δ probed with anti-YlInp1p antibodies. Strains were incubated in YPBO for 9 h. Arrowheads point to a nonspecific immunoreactive polypeptide present in the lysates of both E122 and Ylinp1Δ cells. (C) YlInp1p localizes to the 20KgP subcellular fraction enriched for peroxisomes. Immunoblot analysis of equivalent portions of the 20KgS and 20KgP subcellular fractions from wild-type E122 cells was performed with antibodies to YlInp1p and to the peroxisomal matrix enzyme thiolase (Pot1p). (D) YlInp1p cofractionates with peroxisomes. Organelles in the 20KgP fraction were separated by isopycnic centrifugation on a discontinuous sucrose gradient. Fractions were collected from the bottom of the gradient, and equal portions of each fraction were analyzed by immunoblotting. Fractions enriched for peroxisomes and mitochondria were identified by immunodetection of Pot1p and Sdh2p, respectively. (E) Purified peroxisomes were ruptured by treatment with Ti8 buffer and subjected to ultracentrifugation to obtain a supernatant fraction, Ti8S, enriched for matrix proteins and a pellet fraction, Ti8P, enriched for membrane proteins. The Ti8P fraction was treated further with alkali Na2CO3 and separated by ultracentrifugation into a supernatant fraction (CO3S) enriched for peripherally associated membrane proteins and a pellet fraction (CO3P) enriched for integral membrane proteins. Equivalent portions of each fraction were analyzed by immunoblotting. Immunodetection of Pot1p and Pex2p marked the fractionation profiles of a peroxisomal matrix and integral membrane protein, respectively.
Antibodies raised against YlInp1p specifically recognize an ∼80-kDa polypeptide in whole-cell lysates prepared from wild-type strain E122 but not from the Ylinp1Δ strain (Fig. 5B). Why YlInp1p exhibits a difference between its predicted molecular mass of ∼60 kDa and its ∼80-kDa molecular mass determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis remains unknown.
Subcellular fractionation also showed YlInp1p to be peroxisomal. Similar to the peroxisomal protein thiolase (Pot1p), YlInp1p preferentially localized to a 20,000 × g (20KgP) fraction enriched for peroxisomes and mitochondria (Fig. 5C). Isopycnic density gradient centrifugation of the 20KgP fraction indicated that YlInp1p coenriched with Pot1p and not with the mitochondrial protein Sdh2p (Fig. 5D).
Organelle extraction was used to determine the intraperoxisomal location of YlInp1p (Fig. 5E). Peroxisomes were subjected to hypotonic lysis in dilute alkali Tris buffer, followed by ultracentrifugation to yield a supernatant (Ti8S) fraction enriched for matrix proteins and a pellet (Ti8P) fraction enriched for membrane proteins. Similarly to the peroxisomal integral membrane protein Pex2p, YlInp1p preferentially localized to the Ti8P fraction and not to the Ti8S fraction, like the matrix protein Pot1p. The Ti8P was then extracted with alkali Na2CO3 and subjected to ultracentrifugation. YlInp1p fractionated to the supernatant (CO3S) fraction, consistent with it being a peripheral membrane protein, and not to the pellet (CO3P) fraction, as did Pex2p (Fig. 5E).
Peroxisome inheritance is impaired in cells lacking or overexpressing YlINP1.
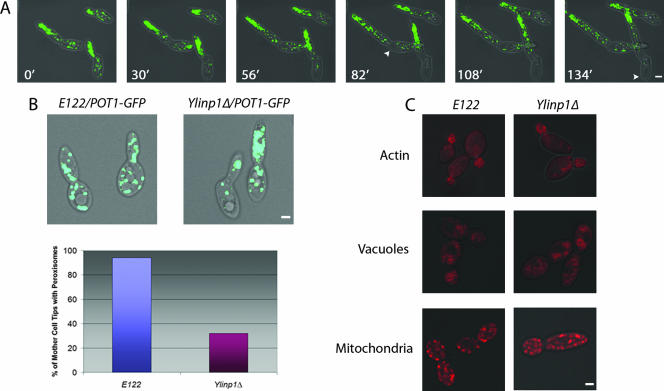
In wild-type Y. lipolytica cells, peroxisomes are essentially evenly distributed between mother cell and bud (Fig. 1A). In contrast, in Ylinp1Δ cells lacking the YlINP1 gene, most peroxisomes are not retained within the mother cell and are transferred to the bud, where they tend to cluster (Fig. 6A; see also video S4 in the supplemental material). Mother cells without peroxisomes are rarely observed in the Ylinp1Δ strain, which contrasts with the situation in S. cerevisiae, where most inp1Δ mother cells fail to retain peroxisomes (5). The small number of peroxisomes remaining in Ylinp1Δ mother cells is not evenly distributed, as peroxisomes in these mother cells prefer to localize near the bud-neck region, thereby leaving the tip region distal to the bud without peroxisomes (Fig. 6B). Quantification showed that 94% of wild-type mother cells retained peroxisomes at their tips distal to the bud, in contrast to only 32% of Ylinp1Δ mother cells (Fig. 6B).
FIG. 6.
Deletion of the YlINP1 gene affects specifically peroxisome inheritance. (A) Peroxisomes of the Ylinp1Δ strain were fluorescently labeled with genomically encoded Pot1p-GFP. Cells were grown for 16 h in YPD medium, transferred to YPBO medium for 16 h, and then visualized at room temperature with an LSM 510 META confocal microscope specially modified for 4D in vivo video microscopy (see Materials and Methods). Representative frames from video S4 in the supplemental material show the specific movements of peroxisomes in the Ylinp1Δ strain. At the start of the video (0′), cells already exhibit pseudohyphal characteristics, and peroxisomes are observed in mother cells and preferentially in buds. The cells at left show bidirectional growth. Continued video imaging showed that peroxisomes continue to move from mother cells to buds and localize to bud tips opposite mother cells (30′ to 133′). Mother cells are largely, but not completely, devoid of peroxisomes. Arrowheads point to tips of mother cells distal to the site of bud emergence that are devoid of peroxisomes. Bar, 2 μm. (B) The wild-type strain E122 and the deletion strain Ylinp1Δ expressing genomically integrated POT1-GFP were grown for 16 h in YPD medium and transferred to YPBO medium for 16 h. Fluorescence images were captured by confocal microscopy. Peroxisome inheritance was quantified as the percentage of mother cells retaining peroxisomes at their tips distal to the site of bud emergence. (C) Deletion of the YlINP1 gene does not affect actin structure or the inheritance of organelles other than peroxisomes. Wild-type E122 and Ylinp1Δ cells synthesizing Pot1p-GFP were grown in YPD medium. Actin was stained with rhodamine-phalloidin, vacuoles were stained with the fluorophore FM4-64, and mitochondria were stained with MitoTracker. Images were captured by confocal microscopy. Bar, 2 μm.
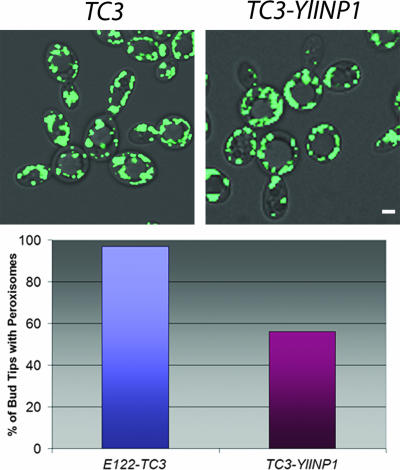
The number of peroxisomes transferred from mother cell to bud is greatly reduced in cells overproducing YlInp1p (Fig. 7). Only 56% of cells overproducing YlInp1p contained peroxisomes at the bud tips. In contrast, 97% of the wild-type cells had peroxisomes at the bud tips (Fig. 7).
FIG. 7.
YlINP1 overexpression leads to peroxisome retention in mother cells. The strain E122/POT1-GFP was transformed with the empty plasmid pTC3 or with pTC3 containing the YlINP1 gene for overexpression of YlINP1. Cells were grown in YND medium and then transferred to and incubated in oleic acid-containing YNO medium for 16 h. Images were captured by confocal microscopy. Quantification of peroxisome retention by mother cells is reported as the percentage of bud tips containing peroxisomes. Bar, 2 μm.
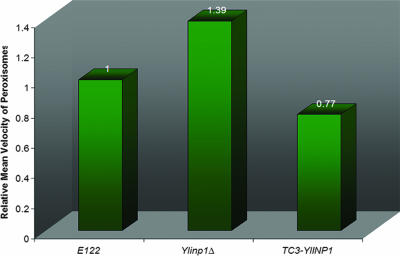
Calculation of the mean velocities of peroxisomes showed that peroxisomes in Ylinp1Δ cells are on average more mobile than those of wild-type cells. In contrast, peroxisomes in cells overexpressing YlINP1 are less mobile than those of wild-type cells (Fig. 8; see also videos S5, S6, and S7 in the supplemental material).
FIG. 8.
Deletion or overexpression of the YlINP1 gene affects peroxisome mobility. The mean velocities of peroxisomes in wild-type E122/POT1-GFP, Ylinp1Δ/POT1-GFP, and YlINP1-overexpressing (TC3-YlINP1/POT1-GFP) cells were determined as described in the legend to Fig. 3.
The structure of actin and the inheritance of other organelles are unaffected in Ylinp1Δ cells.
The impairment in peroxisome inheritance in Ylinp1Δ cells could be due theoretically to some pleiotropic effect resulting from a compromised actin cytoskeleton. We analyzed the organization of the actin cytoskeleton in wild-type cells and cells lacking YlInp1p by staining with rhodamine-phalloidin (Fig. 6C). Actin organization is unchanged in Ylinp1Δ cells.
To test if YlInp1p is required specifically for peroxisome inheritance, we compared the partitioning of other organelles in wild-type and Ylinp1Δ cells. The inheritance of both vacuoles and mitochondria was unimpaired in Ylinp1Δ cells (Fig. 6C).
Moreover, Ylinp1Δ cells did not exhibit a growth defect in YPD medium (unpublished data), suggesting that YlInp1p is not required for the polarized distribution of secretory vesicles.
Ylinp1Δ cells readily undergo the dimorphic transition from yeast to hyphal form.
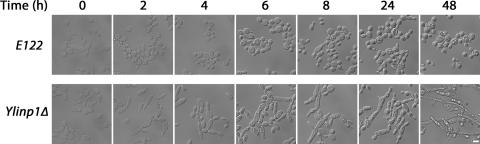
Yeast strains compromised in peroxisome biogenesis usually demonstrate a reduced ability to use oleic acid as the sole source of carbon. Ylinp1Δ cells did not exhibit an overall growth defect on oleic acid-containing YPBO agar medium compared to the wild-type strain (unpublished data). However, Ylinp1Δ cells did show altered cell morphology compared to wild-type cells under the same conditions. Y. lipolytica undergoes a developmentally regulated dimorphic transition from the yeast form to the mycelial form (17). No dimorphic transition was observed in wild-type E122 or Ylinp1Δ cells immediately upon transfer from YPD to YPBO medium (Fig. 9, 0h). E122 cells did not undergo substantial conversion to the hyphal form even after 48 h of incubation in YPBO medium (Fig. 9). In contrast, Ylinp1Δ cells already began to show evidence of the dimorphic transition by 2 h of incubation in YPBO, and all Ylinp1Δ cells were pseudohyphal or hyphal in form by 24 h to 48 h incubation in YPBO medium (Fig. 9). Interestingly, YlInp1p may have additional roles in controlling cell shape and polarity, as Ylinp1Δ cells often exhibit increased width and large ovoid profiles upon continued incubation in YPBO medium (Fig. 9).
FIG. 9.
Deletion of the YlINP1 gene affects the dimorphic transition. Wild-type E122 and Ylinp1Δ cells were grown in YPD medium for 16 h and then transferred to and incubated in YPBO medium. Samples were removed from YPBO at the times indicated and visualized by microscopy. Bar, 5 μm.
DISCUSSION
The distribution of organelles has to be closely controlled during cell division to ensure their faithful segregation between the two resulting cells. Eukaryotic cells that divide by fission usually ensure the accurate inheritance of their organelles by evenly distributing them in the mitotic cell cytoplasm. The cytokinetic machinery that divides the cell into two equally sized daughter cells would thus apportion the organelles evenly between the resultant cells (4, 31). In contrast to cells that divide by median fission, budding yeast must actively and vectorially deliver half of its organelles to the growing bud, while retaining the remaining organelles in the mother cell (18). This feature makes budding yeast more amenable to studies of organelle inheritance, since it facilitates the molecular dissection of organelle inheritance into distinct processes, such as retention of organelles in the mother cell, transport of organelles to the daughter cell, and retention of delivered organelles within daughter cells.
Much progress in our understanding of how peroxisomes partition at cell division has come from studies of the budding yeast S. cerevisiae. In S. cerevisiae, peroxisome dynamics follows a well-defined sequence of events during the cell cycle (3-5, 7). Most peroxisomes are immobilized at the cell periphery, a process dependent on the peroxisomal membrane protein Inp1p (5). During bud growth, half of the maternal peroxisomes are recruited one by one from their static cortical positions and are transported to the bud. The movement of peroxisomes is powered by the class V myosin Myo2p, which is recruited to the peroxisomal membrane via its peroxisome-specific receptor, Inp2p (3, 7). In the bud, Myo2p remains initially attached to Inp2p, which results in the majority of peroxisomes being localized at sites of growth, where Myo2p normally accumulates. Even at cytokinesis, a few peroxisomes in the bud are still engaged by Myo2p and are thus relocated to the mother bud-neck region, whereas the rest remain anchored at the bud cortex, in preparation for the ensuing cell cycle (3).
In the present study, we analyzed peroxisome dynamics in the dimorphic yeast Y. lipolytica. As in S. cerevisiae, most peroxisomes in Y. lipolytica are anchored at the cell periphery. Half of these anchored peroxisomes are then dislodged one at a time from their static positions and transported to the daughter cell. We have shown that peroxisome motility in Y. lipolytica is dependent on the actin cytoskeleton. It would be interesting to determine whether Y. lipolytica peroxisomes engage a myosin motor for their movement or, instead, use the propulsion generated by actin polymerization to advance toward the bud.
We are interested in identifying molecular players implicated in peroxisome inheritance in Y. lipolytica. A search of Y. lipolytica protein databases retrieved one protein of unknown function encoded by the open reading frame YALI0F31229g that exhibits sequence similarity to S. cerevisiae Inp1p. We have designated this protein as YlInp1p. We have shown YlInp1p to be a peripheral membrane protein of peroxisomes involved in peroxisome inheritance. In cells lacking YlInp1p, most peroxisomes were transferred to the bud, with only a few being left in the mother cell. In contrast, in cells overexpressing YlInp1p, peroxisomes are preferentially retained in the mother cell, resulting in buds almost devoid of peroxisomes. These imbalances in peroxisome inheritance resemble the ones observed in S. cerevisiae cells either lacking or overproducing Inp1p, respectively. However, the phenotypes observed in Y. lipolytica strains are not as severe as the ones displayed by the corresponding S. cerevisiae strains. For example, we rarely observed mother cells totally lacking peroxisomes in Ylinp1Δ cells or buds completely devoid of peroxisome in cells overexpressing YlInp1p, as was observed in S. cerevisiae (3, 5). We offer three possible explanations for why Y. lipolytica strains display milder phenotypes than their corresponding S. cerevisiae strains. First, on average, Y. lipolytica contains more peroxisomes per cell than does S. cerevisiae (30 to 40 peroxisomes per cell for Y. lipolytica, as opposed to approximately 10 for S. cerevisiae), which makes the attainment of an extreme phenotype less probable in Y. lipolytica than in S. cerevisiae. Second, other as-yet-unidentified peroxisomal proteins might function in a manner similar to YlInp1p to promote the retention of peroxisomes. This potential functional redundancy would preclude the development of a more dramatic phenotype in cells lacking YlInp1p alone. Third, the de novo synthesis of peroxisomes at the endoplasmic reticulum may be a more rapid process in Y. lipolytica than it is in S. cerevisiae. The production of new peroxisomes would tend to mitigate the imbalances caused by the lack or overproduction of YlInp1p and thus help to alleviate the corresponding phenotypes in Y. lipolytica compared to S. cerevisiae.
Our observations support a role for YlInp1p in peroxisome retention, as previously suggested for S. cerevisiae Inp1p. Most probably, YlInp1p functions as a link between peroxisomes and an anchoring cortical structure. As expected for a protein that would link peroxisomes to an anchoring cortical structure, deletion of the YlINP1 gene leads to peroxisomes that are more mobile than those of wild-type cells, while overexpression of YlINP1 leads to peroxisomes that are largely localized to the cortex of cells and less mobile than peroxisomes of wild-type cells. Even though the existence of anchoring devices suited to retain various organelles in the mother cell has long been proposed, their molecular composition has remained undetermined. Interestingly, retention of mitochondria within S. cerevisiae mother cells has been shown to be dependent on the actin cytoskeleton (37). However, actin patches did not colocalize with cortically immobilized peroxisomes of Y. lipolytica. Moreover, the treatment of wild-type Y. lipolytica cells with the actin-disrupting toxin latrunculin A did not result in the detachment of cortical peroxisomes from their static locations. Taken together, these findings suggest that actin is not involved in the retention of peroxisomes at the cell cortex in Y. lipolytica.
Y. lipolytica is a dimorphic fungus that is able to alternate between a unicellular yeast form and a distinct mycelial form (hyphae and pseudohyphae). Interestingly, in contrast to wild-type cells, Ylinp1Δ cells were observed to undergo substantial conversion from the yeast to the hyphal form when grown in medium containing oleic acid as the sole available carbon source. Usually, filamentous growth is an adaptive strategy employed by nonmotile microorganisms to forage through the environment for scarce nutrients. By restricting growth to the filament tip, cells are able to probe a large volume without investing in a great body mass (9). Cell type switching in dimorphic fungi is known to be modulated by environmental factors, such as nutrient availability (9, 19). Why do cells lacking YlInp1p readily undergo a dimorphic transition to hyphae when grown under conditions requiring peroxisomes? One possibility is that Ylinp1Δ cells are inefficient in metabolizing fatty acids and thus perceive the availability of fatty acids as the sole carbon source as a state of nutrient deprivation. YlInp1p is not required for peroxisome assembly, as Ylinp1Δ cells contain peroxisomes by microscopic analysis. Moreover, these peroxisomes are functional, due to their ability to import proteins targeted by either the PTS1 or the PTS2 signal (unpublished data). However, YlInp1p regulates peroxisome dynamics, which serves a dual purpose. First, it allows peroxisomes to assume correct positioning during cell division, which is required to endow both resulting cells with an equitable number of peroxisomes. Second, it is needed to disperse peroxisomes within cells, thereby increasing their metabolic efficiency (36). Lack of YlInp1p affects the segregation of peroxisomes both within and between cells. In cells lacking YlInp1p, peroxisomes are clustered at the bud tip, leaving other parts of the budded cell almost devoid of peroxisomes. This accumulation of the majority of the peroxisome population at a unique location within cells is likely to result in a decrease in the efficiency of peroxisomal functions. Moreover, the uneven distribution of peroxisomes in cells lacking YlInp1p is likely to trigger the production of new peroxisomes by de novo synthesis at the endoplasmic reticulum. It has been shown that most of the components essential for peroxisome biogenesis in Y. lipolytica are also required for the dimorphic transition from the yeast to the mycelial form and for the delivery of mycelial-form-specific proteins to the plasma membrane (28). Thus, activation of the peroxisome manufacturing machinery might also result in a drastic effect on cell morphology. Collectively, these observations argue that YlInp1p plays an indirect role in regulating dimorphism through its regulation of peroxisome distribution. However, at this time, a direct effect of YlInp1p on cell morphogenesis cannot be excluded.
In closing, we have presented evidence demonstrating that the peroxisomal peripheral membrane protein YlInp1p is directly involved in the inheritance of peroxisomes in Y. lipolytica. YlInp1p probably functions as a peroxisome retention factor, tethering peroxisomes to putative anchoring structures that line the cell periphery (5). To our knowledge, this is the first study of organelle inheritance in a dimorphic yeast.
Acknowledgments
We thank Richard Poirier, Elena Savidov, Hanna Kroliczak, and Dwayne Weber for technical assistance, Monica Fagarasanu for help in preparing figures, Xuejun Sun for help with 4D video microscopy, and members of the Rachubinski laboratory for helpful discussion.
This work was supported by grant 9208 from the Canadian Institutes of Health Research to R.A.R. R.A.R. holds the Canada Research Chair in Cell Biology and is an International Research Scholar of the Howard Hughes Medical Institute. A.F. is the recipient of a Studentship from the Alberta Heritage Foundation for Medical Research.
Footnotes
Published ahead of print on 20 July 2007.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Davidson, R. C., J. R. Blankership, P. R. Kraus, M. de Jesus Berrios, C. M. Hull, C. D'Souza, P. Wang, and J. Heitman. 2002. A PCR-based strategy to generate integrative targeting alleles with large regions of homology. Microbiology 148:2607-2615. [DOI] [PubMed] [Google Scholar]
- 2.de Duve, C., and P. Baudhuin. 1966. Peroxisomes (microbodies and related particles). Physiol. Rev. 46:323-357. [DOI] [PubMed] [Google Scholar]
- 3.Fagarasanu, A., M. Fagarasanu, G. A. Eitzen, J. D. Aitchison, and R. A. Rachubinski. 2006. The peroxisomal membrane protein Inp2p is the peroxisome-specific receptor for the myosin V motor Myo2p of Saccharomyces cerevisiae. Dev. Cell 10:587-600. [DOI] [PubMed] [Google Scholar]
- 4.Fagarasanu, M., A. Fagarasanu, and R. A. Rachubinski. 2006. Sharing the wealth: peroxisome inheritance in budding yeast. Biochim. Biophys. Acta 1763:1669-1677. [DOI] [PubMed] [Google Scholar]
- 5.Fagarasanu, M., A. Fagarasanu, Y. Y. C. Tam, J. D. Aitchison, and R. A. Rachubinski. 2005. Inp1p is a peroxisomal membrane protein required for peroxisome inheritance in Saccharomyces cerevisiae. J. Cell Biol. 169:765-775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gausmann, U., E. Franzl, and C. Kurischko. 1999. Distribution of the actin cytoskeleton during the cell cycle of Yarrowia lipolytica and the visualization of the tubulin cytoskeleton by immunofluorescence. Yeast 15:1079-1086. [DOI] [PubMed] [Google Scholar]
- 7.Hoepfner, D., M. van den Berg, P. Philippsen, H. F. Tabak, and E. H. Hettema. 2001. A role for Vps1p, actin, and the Myo2p motor in peroxisome abundance and inheritance in Saccharomyces cerevisiae. J. Cell Biol. 155:979-990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jansen, G. A., and R. J. A. Wanders. 2006. Alpha-oxidation. Biochim. Biophys. Acta 1763:1403-1412. [DOI] [PubMed] [Google Scholar]
- 9.Kron, S. J. 1997. Filamentous growth in budding yeast. Trends Microbiol. 5:450-454. [DOI] [PubMed] [Google Scholar]
- 10.Kunau, W. H. 2005. Peroxisome biogenesis: end of the debate. Curr. Biol. 15:R774-R776. [DOI] [PubMed] [Google Scholar]
- 11.Lazarow, P. B., and Y. Fujiki. 1985. Biogenesis of peroxisomes. Annu. Rev. Cell Biol. 1:489-530. [DOI] [PubMed] [Google Scholar]
- 12.Lin, Y., L. Sun, L. V. Nguyen, R. A. Rachubinski, and H. M. Goodman. 1999. The Pex16p homologue SSE1 and storage organelle formation in Arabidopsis seeds. Science 284:328-330. [DOI] [PubMed] [Google Scholar]
- 13.Madzak, C., C. Gaillardin, and J.-M. Beckerich. 2004. Heterologous protein expression and secretion in the non-conventional yeast Yarrowia lipolytica: a review. J. Biotechnol. 109:63-81. [DOI] [PubMed] [Google Scholar]
- 14.Mullen, R. T., and R. N. Trelease. 2006. The ER-peroxisome connection in plants: development of the “ER semi-autonomous peroxisome maturation and replication” model for plant peroxisome biogenesis. Biochim. Biophys. Acta 1763:1655-1668. [DOI] [PubMed] [Google Scholar]
- 15.Poirier, Y., V. D. Antonenkov, T. Glumoff, and J. K. Hiltunen. 2006. Peroxisomal β-oxidation: a metabolic pathway with multiple functions. Biochim. Biophys. Acta 1763:1413-1426. [DOI] [PubMed] [Google Scholar]
- 16.Pruyne, D., A. Legesse-Miller, L. Gao, Y. Dong, and A. Bretscher. 2004. Mechanisms of polarized growth and organelle segregation in yeast. Annu. Rev. Cell Dev. Biol. 20:559-591. [DOI] [PubMed] [Google Scholar]
- 17.Rodriguez, C., and A. Dominguez. 1984. The growth characteristics of Saccharomycopsis lipolytica: morphology and induction of mycelium formation. Can. J. Microbiol. 30:605-612. [Google Scholar]
- 18.Rossanese, O. W., C. A. Reinke, B. J. Bevis, A. T. Hammond, I. B. Sears, J. O'Connor, and B. S. Glick. 2001. A role for actin, Cdc1p, and Myo2p in the inheritance of late Golgi elements in Saccharomyces cerevisiae. J. Cell Biol. 153:47-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanchez-Martinez, C., and J. Perez-Martin. 2001. Dimorphism in fungal pathogens: Candida albicans and Ustilago maydis—similar inputs, different outputs. Curr. Opin. Microbiol. 4:214-221. [DOI] [PubMed] [Google Scholar]
- 20.Smith, J. J., M. Marelli, R. H. Christmas, F. J. Vizeacoumar, D. J. Dilworth, T. Ideker, T. Galitski, K. Dimitrov, R. A. Rachubinski, and J. D. Aitchison. 2002. Transcriptome profiling to identify genes involved in peroxisome assembly and function. J. Cell Biol. 158:259-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Szilard, R. K., V. I. Titorenko, M. Veenhuis, and R. A. Rachubinski. 1995. Pay32p of the yeast Yarrowia lipolytica is an intraperoxisomal component of the matrix protein translocation machinery. J. Cell Biol. 131:1453-1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tabak, H. F., D. Hoepfner, A. van der Zand, H. J. Geuze, I. Braakman, and M. A. Huynen. 2006. Formation of peroxisomes: present and past. Biochim. Biophys. Acta 1763:1647-1654. [DOI] [PubMed] [Google Scholar]
- 23.Tam, Y. Y. C., A. Fagarasanu, M. Fagarasanu, and R. A. Rachubinski. 2005. Pex3p initiates the formation of a preperoxisomal compartment from a subdomain of the endoplasmic reticulum in Saccharomyces cerevisiae. J. Biol. Chem. 280:34933-34939. [DOI] [PubMed] [Google Scholar]
- 24.Tam, Y. Y. C., J. C. Torres-Guzman, F. J. Vizeacoumar, J. J. Smith, M. Marelli, J. D. Aitchison, and R. A. Rachubinski. 2003. Pex11-related proteins in peroxisome dynamics: a role for the novel peroxin Pex27p in controlling peroxisome size and number. Mol. Biol. Cell 14:4089-4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thoms, S., and R. Erdmann. 2005. Dynamin-related proteins and Pex11 proteins in peroxisome division and proliferation. FEBS J. 272:5169-5181. [DOI] [PubMed] [Google Scholar]
- 26.Titorenko, V. I., and R. T. Mullen. 2006. Peroxisome biogenesis: the peroxisomal endomembrane system and the role of the ER. J. Cell Biol. 174:11-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Titorenko, V. I., G. A. Eitzen, and R. A. Rachubinski. 1996. Mutations in the PAY5 gene of the yeast Yarrowia lipolytica cause the accumulation of multiple subpopulations of peroxisomes. J. Biol. Chem. 271:20307-20314. [DOI] [PubMed] [Google Scholar]
- 28.Titorenko, V. I., D. M. Ogrydziak, and R. A. Rachubinski. 1997. Four distinct secretory pathways serve protein secretion, cell surface growth, and peroxisome biogenesis in the yeast Yarrowia lipolytica. Mol. Cell. Biol. 17:5210-5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van den Bosch, H., R. B. H. Schutgens, R. J. A. Wanders, and J. M. Tager. 1992. Biochemistry of peroxisomes. Annu. Rev. Biochem. 61:157-197. [DOI] [PubMed] [Google Scholar]
- 30.van der Klei, I. J., and M. Veenhuis. 2006. Yeast and filamentous fungi as model organisms in microbody research. Biochim. Biophys. Acta 1763:1364-1373. [DOI] [PubMed] [Google Scholar]
- 31.Warren, G., and W. Wickner. 1996. Organelle inheritance. Cell 84:395-400. [DOI] [PubMed] [Google Scholar]
- 32.Weisman, L. S. 2003. Yeast vacuole inheritance and dynamics. Annu. Rev. Genet. 37:435-460. [DOI] [PubMed] [Google Scholar]
- 33.Weisman, L. S. 2006. Organelles on the move: insights from yeast vacuole inheritance. Nat. Rev. Mol. Cell Biol. 7:243-252. [DOI] [PubMed] [Google Scholar]
- 34.Xiang, X., and M. Plamann. 2003. Cytoskeleton and motor proteins in filamentous fungi. Curr. Opin. Microbiol. 6:628-633. [DOI] [PubMed] [Google Scholar]
- 35.Yaffe, M. P. 1999. The machinery of mitochondrial inheritance and behavior. Science 283:1493-1497. [DOI] [PubMed] [Google Scholar]
- 36.Yan, M., N. Rayapuram, and S. Subramani. 2005. The control of peroxisome number and size during division and proliferation. Curr. Opin. Cell Biol. 17:376-383. [DOI] [PubMed] [Google Scholar]
- 37.Yang, H. C., A. Palazzo, T. C. Swayne, and L. A. Pon. 1999. A retention mechanism for distribution of mitochondria during cell division in budding yeast. Curr. Biol. 9:1111-1114. [DOI] [PubMed] [Google Scholar]