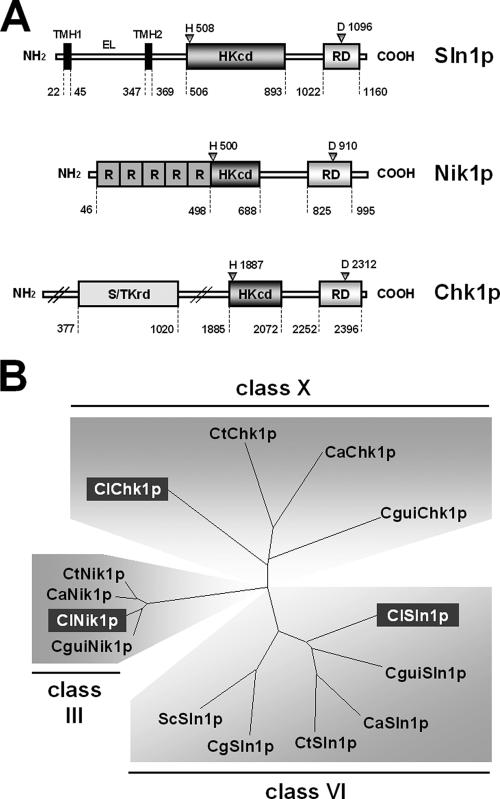
FIG. 1.
(A) Structure of the C. lusitaniae HKR proteins. The putative phosphorylable residues are indicated with triangles. EL, extracellular loop; HKcd, histidine kinase catalytic domain; R, repeated sequence; RD, receiver domain; S/TKrd, serine/threonine kinase-related domain; TMH, transmembrane helix. (B) Dendrogram generated after alignment of the predicted sequences of C. lusitaniae HKRs (ClSln1p, ClNik1p, and ClChk1p) with sequences retrieved from C. guilliermondii (CguiSln1p, CguiNik1p, and CguiChk1p), C. tropicalis (CtSln1p, CtNik1p, and CtChk1p), C. glabrata (CgSln1p), and S. cerevisiae (ScSln1p). Alignment utilizes the neighborhood-joining method from TreeView PPC software. Distances along the branches represent the divergence between two cognate sequences.