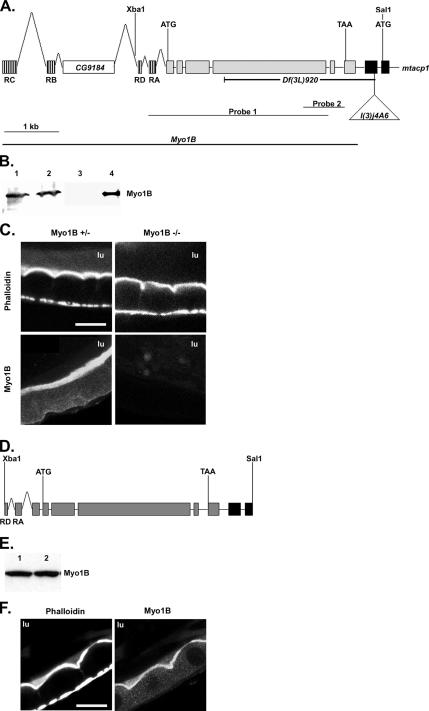
Figure 3.
Creation of a Myo1B deletion mutation. (A) The genomic structure of Myo1B and part of mtacp1 are shown. Transcription start and stop sites are labeled with ATG and TAA, respectively. The position of the selected restriction sites are shown above and the P{lacW}l(3)j4A6 insertion below. The four predicted isoforms of Myo1b use different alternatively spliced first exons, these are shown hashed, only one is used in each allele; the first 2 exons of mtacp1 are filled black. The CG9184 gene is found in the Myo1b intronic sequence. The extent of the Df(3L)920 deficiency is shown below. Probes 1 and 2 represent the Southern probes used to identify deletion mutations in Myo1B. (B) Western blot showing Myo1B expression in P{lacW}I(3)j4A6 and the Df(3L)920 mutation. Lane 1, homozygous P insertion; lane 2, heterozygous P insertion; lane 3, homozygous deletion; lane 4, heterozygous deletion. (C) Myo1B mutant and heterozygous control anterior midgut costained for actin and Myo1B. Loss of Myo1B does not disrupt actin distribution in midgut enterocyte BBs. (D) An Xba1–Sal 1 fragment of genomic DNA was used to rescue the loss of Myo1B in the deletion mutation. This construct contains two of the four alternate first exons. Transcription start and stop sites are represented by ATG and TAA, respectively. (E) Western blot showing expression of P{Myo1B} in the deletion background. Lane 1, P{Myo1B} heterozygote; lane 2, P{Myo1B} homozygote. Myo1B is expressed at equal levels in both genotypes. (F) Anterior midgut of the P{Myo1B} rescue line costained for actin and Myo1B. Transgene-expressed Myo1B localizes to the BB of midgut enterocytes. Bar, 20 μm (C and F). lu, midgut lumen.