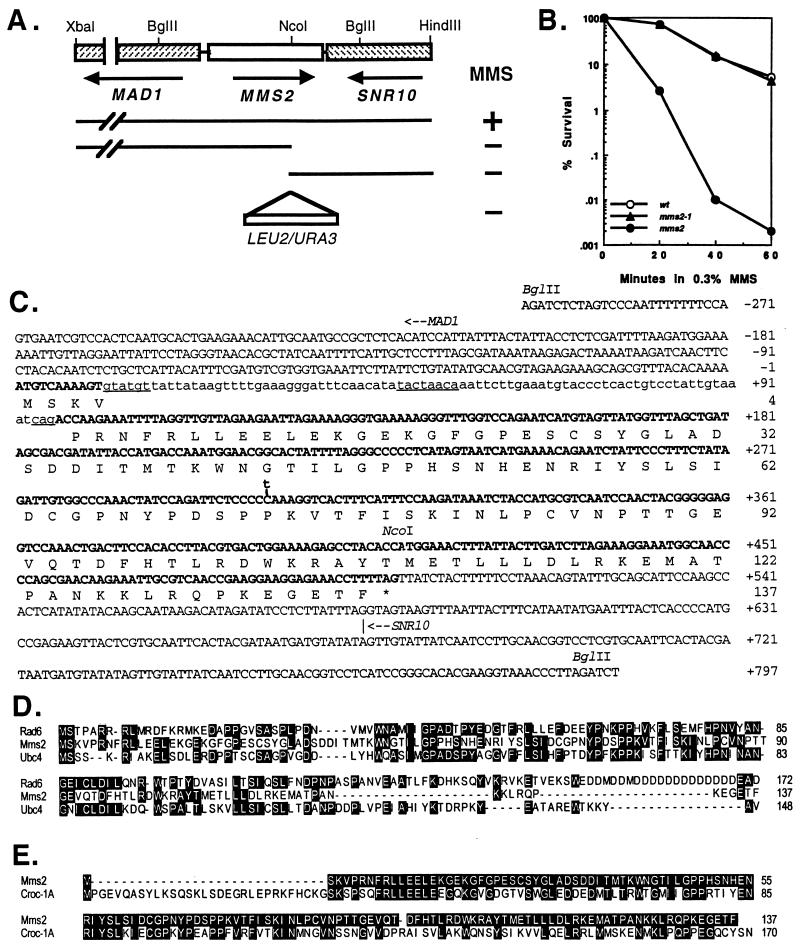
Figure 1.
Physical characterization of the S. cerevisiae MMS2 gene. (A) Mapping and disruption of the MMS2 gene. A subclone containing the 3-kb XbaI–HindIII fragment enables (+) the mms2-1 mutant to grow on YPD plates containing 0.4% MMS. Further deletions to the NcoI site from either end abolished (−) the MMS2 function. Either a URA3 or a LEU2 fragment was inserted at the NcoI site to construct the mms2∷URA3 and mms2∷LEU2 disruption cassettes. (B) Killing of DBY747 (wt), WX17–4a (mms2-1), and SBU (mms2) in a liquid culture containing 0.3% MMS. (C) The nucleotide and deduced amino acid sequences of the MMS2 gene (GenBank accession no. U66724). Exons are in boldface. Lowercase indicates intron sequences. Consensus sequences within the intron are underlined. The translation initiation site for MAD1 and the transcriptional termination site for SNR10 are indicated with an arrow for direction. A C-to-T transition found in the mms2-1 mutation at nucleotide 303 is marked. (D) Amino acid sequence alignments of Mms2 with two yeast Ubc proteins, Rad6 (Ubc2) and Ubc4. Residues shared by two or more proteins are highlighted. (E) Amino acid sequence alignment of Mms2 with Croc1. Residues in Croc1 identical to Mms2 are highlighted.