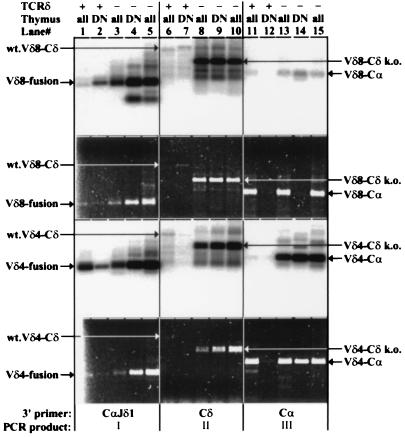
Figure 4.
RT–PCR analysis of VDJδ transcripts from wild-type (TCRδ+) and TCRδ-deficient (TCRδ−) total (all) or sorted HSAhi, CD25+, CD4/CD8 double negative (DN) thymocytes. The first and third panels from the top show hybridization to a Jδ1 gene-specific oligonucleotide probe. The second and fourth panels show the corresponding gels stained with ethidium bromide and visualized with UV illumination. Two independent TCRδ total thymus samples are shown. The top two panels show results obtained with a Vδ8-specific forward primer, and the bottom two show results with a Vδ4-specific forward primer. Lanes: 1–5, PCRs performed with the 3′CαJδ1 hybrid reverse primer; 6–10, PCRs performed with a Cδ reverse primer; 11–15, PCRs performed with a Cα reverse primer. The predominant PCR product (I, II, or III, as defined in Fig. 1) for each group of five lanes is indicated at the bottom. The discrepancy observed between the hybridization and ethidium staining signals for Vδ8 RT–PCR reactions in lanes 1 vs. 2, lanes 4 vs. 5, and lanes 13 and 15 vs. 14 is probably due to mispriming (lanes 1–5) or priming (lanes 13–15) of RT–PCR from VJα-Cα templates abundant in total thymus but absent from DN thymus templates. Note the difference in the size of the Cδ-specific RT–PCR products between wild-type and TCRδ-deficient mice due to the targeted deletion of the Cδ sequences. The identity of these bands was assigned based on their expected sizes and for the Vδ4-Cδ products from TCRδ-deficient samples was also confirmed by DNA sequencing.