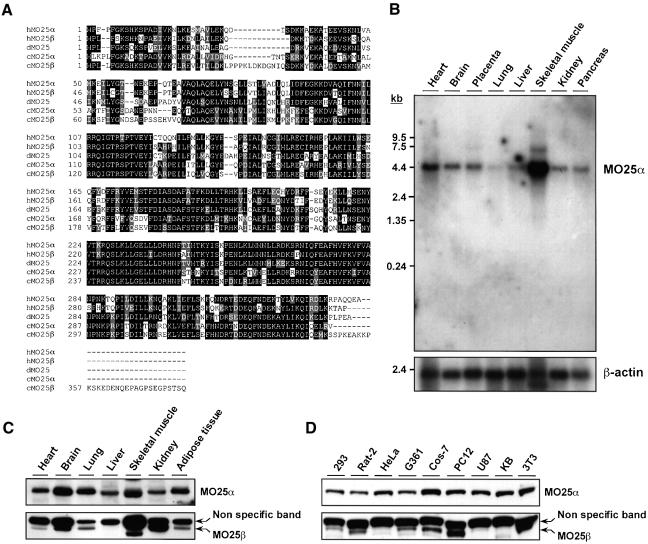
Fig. 2. Amino acid sequence and tissue distribution patterns of MO25α and MO25β isoforms. (A) Amino acid sequence alignment of the human MO25α (NCBI accession No. NP_057373) and MO25β (NCBI accession No. CAC37735) isoforms as well as C.elegans MO25α (NCBI accession No. CAB16486) and MO25β (NCBI accession No. NP_508691) and Drosophila MO25 (NCBI accession No. P91891) putative homologues. Conserved residues are boxed in black, and homologous residues are shaded in grey. Sequence alignments were performed using the CLUSTALW and BOXSHADE programmes at http://www.ch.embnet.org/ using standard parameters. (B) A 32P-labelled fragment of the MO25α cDNA was used to probe a northern blot containing polyadenylated RNA isolated from the indicated human tissues. The membrane was autoradiographed, and the MO25α probe was observed to hybridize to a 4.2-kb message, identical to the size predicted for the MO25α message from database analysis. As a loading control, the northern blot was hybridized with a β-actin probe. (C and D) The indicated mouse tissue (C) or cell (D) extracts containing 20 µg of total cell protein were immunoblotted with anti-MO25α and anti-MO25β antibodies.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.