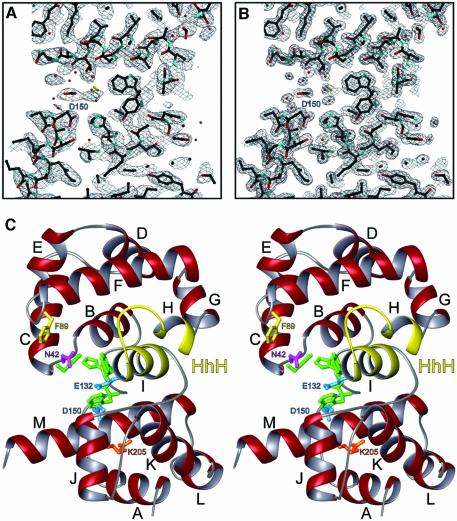
Fig. 1. The structure of MagIII. (A) A cross-section of the refined model and the 2.7 Å solvent-flattened experimental electron density map, contoured at 1σ. (B) Same view of the refined model against the final σA-weighted 2mFo–DFc electron density map contoured at 1σ. (C) Stereoscopic ribbon representation of MagIII with the HhH domain at the top and the N/C-terminal domain at the bottom of the figure. Helices are labeled A–M, and the HhH motif is highlighted yellow. The nucleobase binding residues are colored green and putative catalytic residues are blue. DNA intercalating (Asn42, magenta) and ‘wedge’ (Phe45, yellow) residues responsible for base flipping are also shown, as well as the carbamylated lysine residue in the N/C-terminal domain (orange).

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.