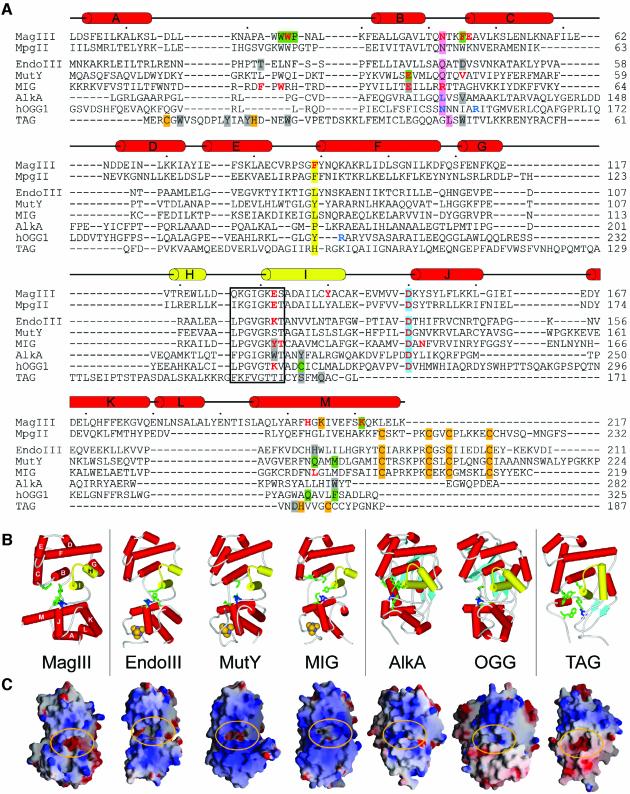
Fig. 2. The HhH superfamily of DNA glycosylases. (A) The structure-based sequence alignment of MagIII with the HhH glycosylase structures and the MpgII sequence was prepared using the program SEQUOIA (Bruns et al., 1999). MagIII secondary structure elements are shown schematically, with the HhH motif highlighted as yellow cylinders (helices H–I of MagIII). The HhH residues that contact the DNA in the AlkA–DNA complex are boxed, and the conserved catalytic aspartic acid is shaded blue. Residues in the nucleobase binding pocket confirmed (green) or predicted (gray) to contact the target base are shaded, and the positions of the side chains that intercalate the DNA helix at the lesioned and non-lesioned strands are shaded pink and yellow, respectively. Residues that contact the orphaned DNA base opposite the modified base in AlkA and Ogg1 are colored blue, while residues shown by mutagenesis to be important for either catalysis or DNA binding are colored red. Side chains that coordinate the Fe4S4 clusters (MpgII, EndoIII, MutY, MIG) and Zn2+ ion (TAG), as well as the carbamylated lysine in MagIII are shaded orange. (B) Schematic representations of the HhH glycosylase structures are oriented as in Figure 1C. Helices are shown as red and yellow (HhH motif) cylinders, β-sheets as light blue arrows, and Fe4S4 clusters as golden CPK spheres. Side chains of functionally significant active site residues are rendered as sticks, with the conserved aspartic acid colored dark blue. (C) Solvent accessible surfaces are colored according to electrostatic potential (blue, positive; red, negative) and were created with GRASP (Nicholls et al., 1991). The substrate binding pockets at the domain interface are circled. The structures have been rotated ∼90° with respect to the views shown in (B). Larger versions of (B) and (C) are available as Supplementary data.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.