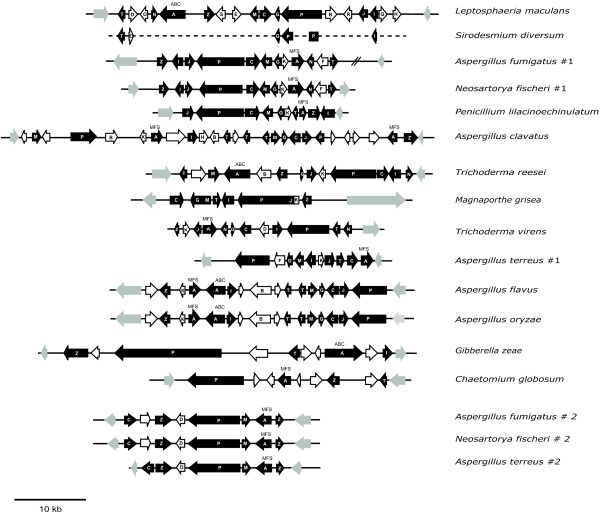
Figure 2.
Putative ETP biosynthetic gene clusters in ascomycetes. Genes (white text on black background) include those with best matches to non-ribosomal peptide synthetase (P), thioredoxin reductase (T), methyl transferases (M and N), glutathione S-transferase (G) and cytochrome P450 monooxygenase (C), ACCS (I), dipeptidase (J), as well as a transcriptional regulator (Z) and a transporter (A – Multi Facilitator Superfamily (MFS) or ABC). In addition genes with predicted roles in modification of the side chains of the core ETP moiety are noted (see Table 1). Genes with no lettering are either hypothetical or have no strong matches to ETP cluster genes. Genes shaded in grey are predicted to flank the cluster and encode proteins with best matches to proteins with no potential roles in ETP biosynthesis. The cluster in T. virens might be incomplete as a gene common to ETP cluster was at one end of a single cosmid clone. In M. grisea G and M, and P, J and K are annotated as fused genes. The arrangement of genes in S. diversum is based on the sirodesmin PL cluster in L. maculans; the dashed line represents unsequenced regions. '#2' after taxon name indicates that this is the second cluster found in that species.