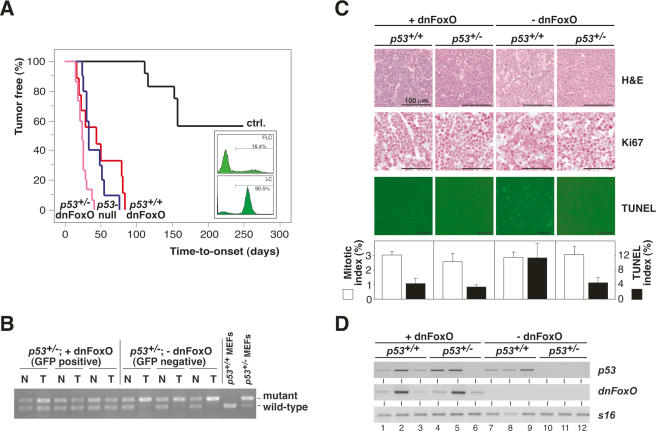
Figure 1.
The dnFoxO moiety accelerates Myc-driven lymphomagenesis by blocking p53-dependent apoptosis. (A) Lymphoma incidence in recipient mice of Eμ-myc; p53+/+ or Eμ-myc; p53+/− hematopoietic stem cells stably transduced with dnFoxO (p53+/+;dnFoxO; n = 9 [red]; and p53+/−;dnFoxO; n = 15 [purple]) or mock-infected (p53+/+;mock [ctrl.]; n = 12 [black]; and p53+/−;mock [p53-null]; n = 10 [blue]). (Insert) Representative flow cytometric GFP scans of Eμ-myc; p53+/+ fetal liver cells (FLC) infected with the MSCV-dnFoxO-IRES-GFP retrovirus (top) and of lymphoma cells (LC) arising from this population after stem cell transplantation (percentage reflects fraction of GFP-positive cells), indicating positive selection of the dnFoxO moiety (bottom). (B) Allele-specific p53 PCR from genomic DNA extracted from representative p53+/−; dnFoxO (+dnFoxO) and p53+/−; mock (−dnFoxO) lymphoma samples and p53+/+ and p53+/− MEFs as internal PCR control (normal tissue [N], lymphoma tissue from the same animal [T]). (C) Lymph node histopathology of the indicated genotypes sampled at lymphoma diagnosis to visualize histomorphology and mitotic figures by hematoxylin/eosin (H&E), proliferation by Ki67, and spontaneous apoptosis by TUNEL staining in situ, and their respective quantifications. (D) Expression analysis of p53 and dnFoxO transcripts by RT–PCR in individual lymphomas (n = 3 per genotype) with S16 as a control.