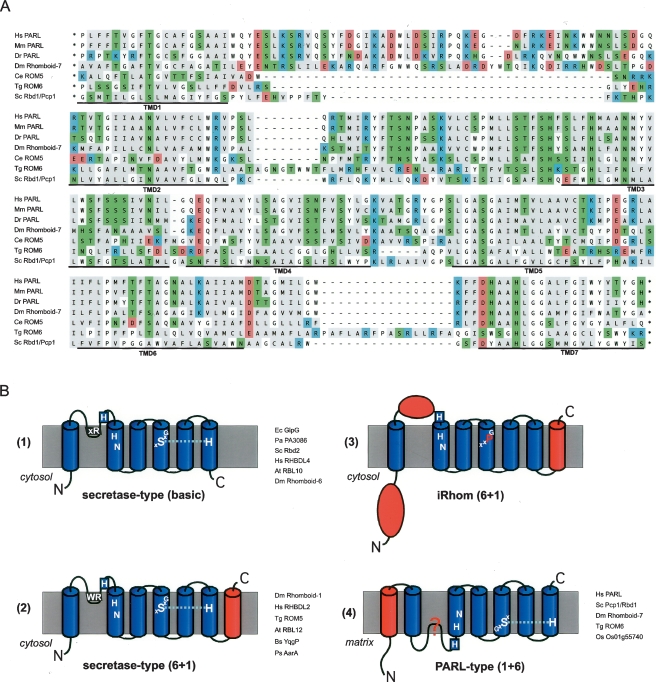
Figure 2.
Rhomboid topology. (A) Multiple-sequence alignment of the membrane integral portion of PARL-type rhomboids from human (Homo sapiens, Hs), mouse (Mus musculus, Mm), zebrafish (Danio rerio, Dr), Drosophila melanogaster (Dm; Rhomboid-7), Caenorhabditis elegans (Ce; named ROM5 by automated annotation), Toxoplasma gondii (Tg; ROM6), and Saccharomyces cerevisiae (Sc; Pcp1/Rbd1). Assuming the overall protein architecture is conserved, we manually corrected gaps in predicted TMDs of the ClustalW-based alignment. Typically for rhomboids, the TMDs have a high content of polar amino acids, which occur predominantly in conserved positions. In the alignment, the functional characteristics of the amino acids are indicated by background color (acidic, red; strong basic, blue; polar and weak basic, green; and hydrophobic, gray). (B) Topology models for different rhomboid proteases and catalytically inert iRhoms; extra domains fused to the basic six TMD rhomboid core are highlighted in red; the key conserved residues and the L1 structure extending sidewise in the membrane are indicated (Urban et al. 2001; Lemberg et al. 2005; Wang et al. 2006); examples of bacterial and eukaryotic rhomboids are listed. (For accession numbers, see Table S1; Figs. 1, 3.)