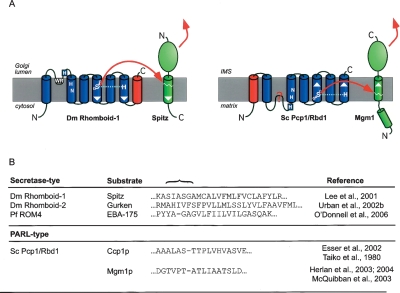
Figure 6.
Secretase- and PARL-type rhomboids have active sites with opposite orientations. (A) Topology model of Drosophila Rhomboid-1 (Urban et al. 2001) and S. cerevisiae PARL (Pcp1/Rbd1). Cleavage by Drosophila Rhomboid-1 releases the N-terminal portion of the membrane tethered growth factor Spitz into the Golgi lumen, thereby allowing its secretion to trigger EGFR signaling in neighboring cells (Lee et al. 2001). In contrast, S. cerevisiae PARL, Pcp1/Rbd1, cleaves its substrate Mgm1 to release the C-terminal portion into the intermembrane space (IMS) (Herlan et al. 2004). Topology model as in Figure 2B; extra TMDs fused to the C terminus of Drosophila Rhomboid-1 and the N terminus of PARL (Pcp1/Rbd1) are colored red. The orientation of the active site TMDs and substrate TMD is indicated by white arrowheads. (B) Examples of substrate TMDs of secretase- and PARL-type rhomboids. Cleavage site region is indicated by a brace. Experimentally determined cleavage sites are indicated in the sequences by dashes. Note that the substrates for S. cerevisiae PARL (Pcp1/Rbd1) are cleaved in secondary non-typical short TMDs (Takio et al. 1980; Herlan et al. 2003, 2004).